Fig. 1.
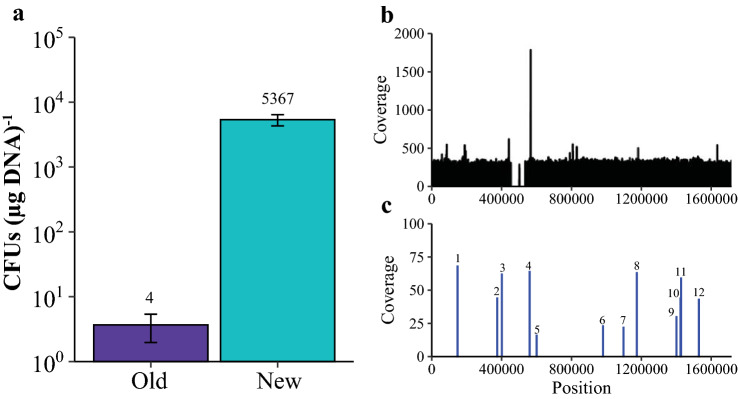
Optimized transposon mutagenesis produces random insertions across genome that can be mapped with whole genome sequencing: A Comparison of methods described by Sattler et al. (Sattler et al. 2013) and Quitzke et al. (Quitzke et al. 2018) [old] and the method described in this study [new]. Data are averages from three independent experiments, and error bars represent one standard deviation around the mean. B Coverage distribution of reads mapped from a representative sequencing run. Coverage data created by Bowtie2 alignment of all forward reads to reference genome of M. maripaludis JJ (CP026606). C Sites of random transposon insertion across genome of 12 randomly picked puromycin grown (containing transposon) colonies. Two Illumina reactions containing 6 random mutants each were sequenced and mapped to identify mini-mariner transposition. Insertion sites position and coverage value were determined through visualization in Geneious prime® (v.2021.0.3). Site determined by presence of AT (known integration site for mariner-based transposition) and where reads of either end of the transposon overlapped