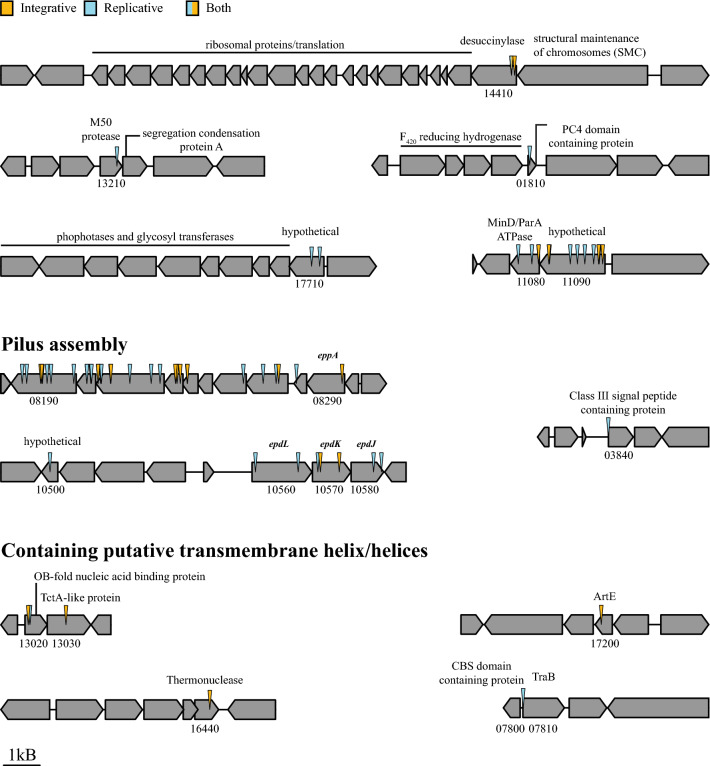
Fig. 2.
Schematic representation of insertions and respective operons: For each candidate mutant, insertion site was determined via visualization in Geneious prime®(v.2021.0.3). Site determined by presence of AT (known integration site for mariner-based transposition) and where reads of either end of the transposon overlapped. Genes around the insertion are included for context. Genes of interest are referenced using the locus tag format of Poehlein et al. (MMJJ_XXXXX) (Poehlein et al. 2018). Select annotations were added manually based on either NCBI or UniProt annotations. Colors used to mark which DNA substrate used in screen where replicative refers to pLW40neo and integrative refers to pCRUptNeoΔflaK. One insertion in MMJJ_14400 was identified in both substrates, therefore it was colored with both. A table summary for each of these insertions is provided as Table 1