Figure 1. Relationship of mesenchymal cell populations across the intestinal length.
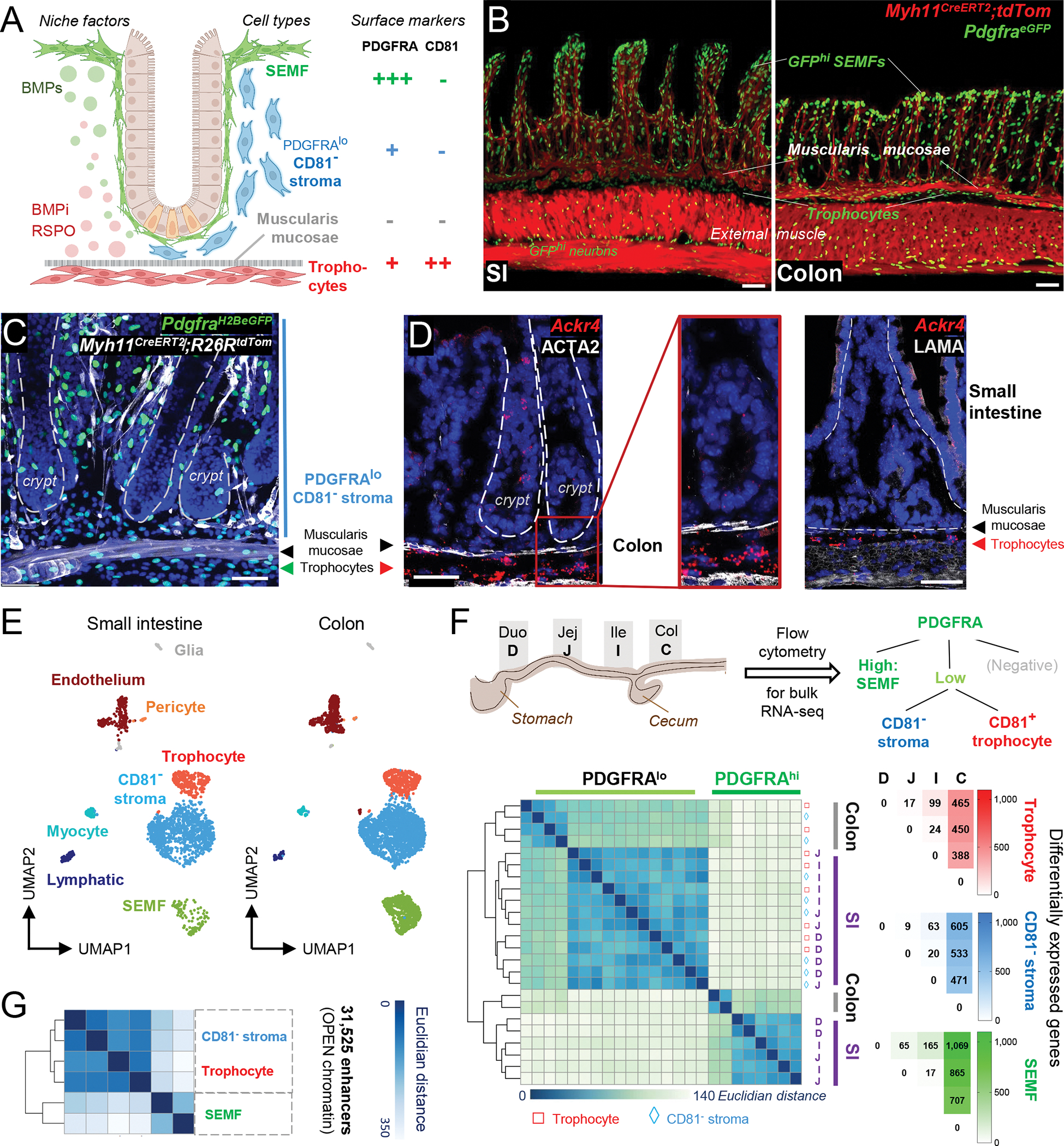
A) Cell populations and signaling in SI crypts. BMPi provided by sub-cryptal trophocytes (red, PDGFRAlo CD81+) oppose agonists provided by dense SEMF (green, PDGFRAhi) aggregates at the crypt-villus junction. The function of supra-muscularis PDGFRAlo cells (blue), which lack CD81 expression, is unknown and is the topic of this study.
B) Confocal images of Myh11CreERT2;R26RLSL-tdTom;PdgfraH2BeGFP mouse ileum (SI, left, 24 μm z-stack) and colon (right, 15 μm z-stack). Scale bars 50 μm.
C) Left, whole-mount confocal micrograph (26-μm z-stack) of colonic tissue from PdgfraH2B-eGFP;Myh11CreERT2;R26RtdTom mice. White: Myh11Cre-marked smooth muscle, green: GFP+ nuclei, blue (DAPI): cell nuclei. Dashed lines demarcate crypts. Scale bar 50 μm.
D) In situ hybridization for trophocyte marker Ackr4 (red) in colonic crypts counterstained with ACTA2 antibody (white). Rare epithelial Ackr4+ cells are tuft cells. Dashed lines demarcate crypts. Scale bars 50 μm.
E) scRNAseq-derived UMAP clustering of unfractionated SI (n=2,042)17 and colonic (n=3,940) EPCAM− CD45− mesenchymal cell types identified using known cell-specific markers (Figure S1B).
F) Differential PDGFRA and CD81 expression allow purification of 3 dominant PDGFRA+ cell types from each intestinal segment: duodenum (D), jejunum (J), ileum (I), and colon (C). Left, Unsupervised hierarchical clustering of duplicate bulk RNA-seq profiles of SEMFs (green), trophocytes (red), and CD81− stromal cells (blue) from each intestinal segment; the latter two populations cluster together. Color scale: Euclidean distance of samples. Right, Numbers of differentially expressed genes (Padj <0.01, log2 fold-difference >|1.5|) in each cell type and intestinal segment.
G) Hierarchical clustering of duplicate PDGFRA+ cell samples from whole SI by similarities in open chromatin reads (ATAC-Seq) at the union set of 31,525 enhancers (> −1 kb, > 2 kb from transcription start sites). Color scale: Euclidean distance.
See also Figure S1.
