FIGURE 4.
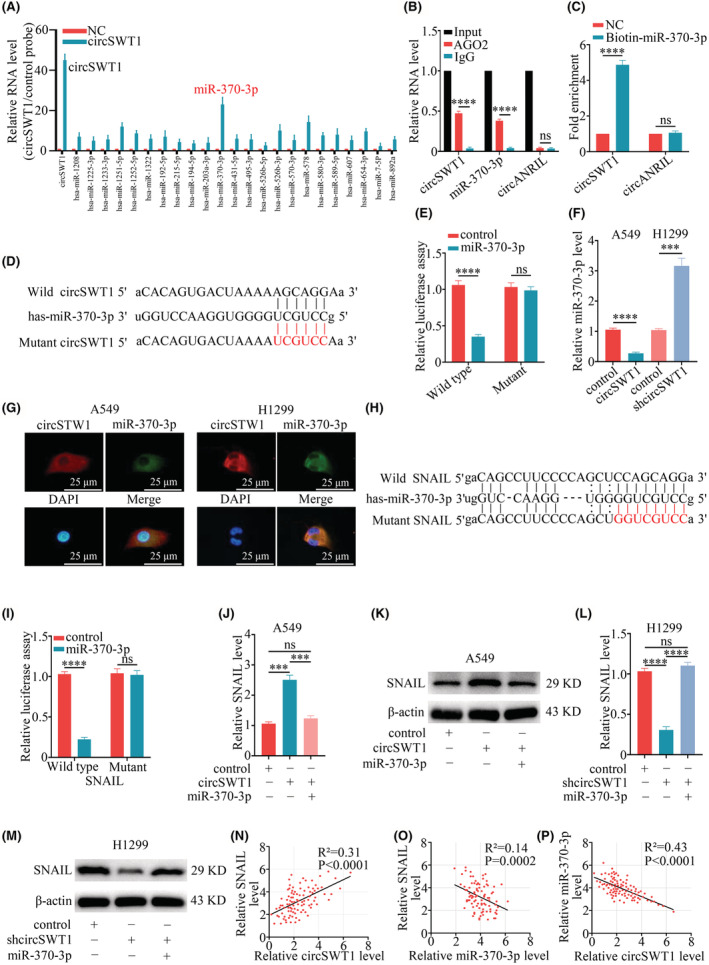
CircSWT1 upregulates the expression of the downstream molecule SNAIL by sponging miR‐370‐3p. (A) The relative RNA level of H1299 cells was measured with a biotinylated circSWT1 probe in the circRIP assay. (B) The RIP assay was used to verify the interaction between circSWT1 and miR‐370‐3p by using the anti‐AGO2 antibody in H1299 cells. (C) The enrichment of circSWT1 in H1299 cells transfected with biotinylated miR‐370‐3p mimics was measured by the RNA pulldown assay, and circANRIL acted as a negative control. (D) The potential binding sites between circSWT1 and miR‐370‐3p. (E) The luciferase activity of wild‐type and mutant circSWT1 in HEK‐293 T cells transfected with miR‐370‐3p mimics. (F) The level of miR‐370‐3p significantly decreased in A549 cells with circSWT1 overexpression and significantly increased in H1299 cells with circSWT1 knockdown compared to the controls. (G) The images of colocalization between circSWT1 and miR‐370‐3p in A549 and H1299 cells via RNA FISH staining were captured with a fluorescence microscope. Nuclei were stained with DAPI. (H) The putative binding sites between SNAIL and miR‐370‐3p. (I) The luciferase activity of SNAIL (wild‐type and mutant type) was measured in HEK‐293 T cells transfected with miR‐370‐3p mimics. (J) The expression of SNAIL was quantified with qRT‐PCR after the dual overexpression of circSWT1 and miR‐370‐3p in A549 cells. (K) The expression of SNAIL was estimated with Western blotting after the dual overexpression of circSWT1 and miR‐370‐3p in A549 cells. (L) The expression of SNAIL was quantified with qRT‐PCR after the dual knockdown of circSWT1 and miR‐370‐3p in H1299 cells. (M) The expression of SNAIL was estimated with Western blotting after the dual knockdown of circSWT1 and miR‐370‐3p in H1299 cells. [(N)–(P)] The correlations among the levels of circSWT1, miR‐370‐3p and SNAIL in 96 NSCLC patients were assessed by qRT–PCR. The data are presented as the mean ± SD of three independent experiments. ***p < 0.001, ****p < 0.0001, ns: not significant.
