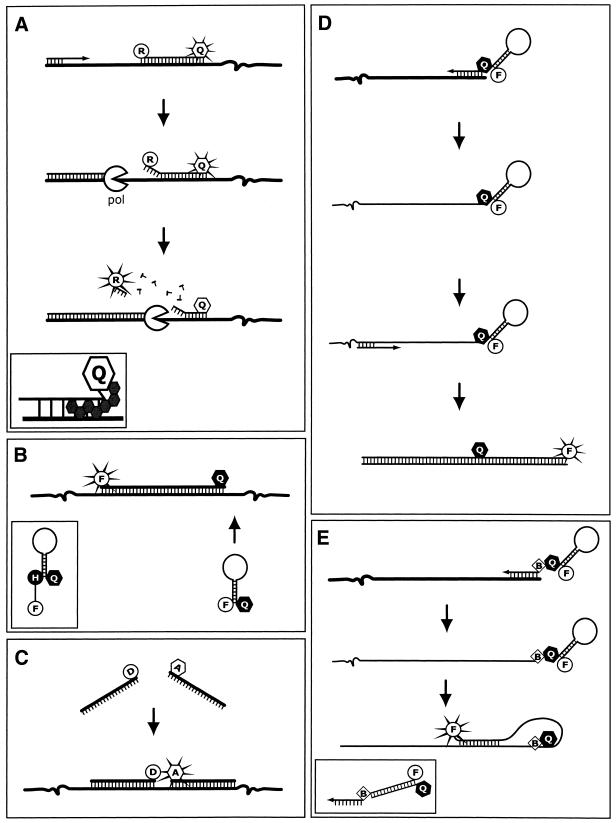
Figure 3.
Oligoprobe chemistries. (A) 5′ Nuclease oligoprobes. As the DNA polymerase (pol) progresses along the relevant strand, it displaces and then hydrolyses the oligoprobe via its 5′→3′ endonuclease activity. Once the reporter (R) is removed from the extinguishing influence of the quencher (Q, open), it is able to release excitation energy at a wavelength that is monitored by the instrument and different from the emissions of the quencher. Inset shows the NFQ and MGB molecule that make up the improved MGB nuclease-oligoprobes. (B) Hairpin oligoprobes. Hybridisation of the oligoprobe to the target separates the fluorophore (F) and non-fluorescent quencher (Q, closed) sufficiently to allow emission from the excited fluorophore, which is monitored. Inset shows a wavelength-shifting hairpin oligoprobe incorporating a harvester molecule. (C) Adjacent oligoprobes. Adjacent hybridisation results in a FRET signal due to interaction between the donor (D) and acceptor (A) fluorophores. This bimolecular system acquires its data from the acceptor’s emissions in an opposite manner to the function of nuclease oligoprobe chemistry. (D) Sunrise primers. The opposite strand is duplicated so that the primer’s hairpin structure can be disrupted. This separates the labels, eliminating the quenching in a similar manner to the hairpin oligoprobe. (E) Scorpion primers. The primer does not require extension of the complementary strand; in fact it blocks extension to ensure that the hairpin in the probe is only disrupted by specific hybridisation with a complementary sequence designed to occur downstream of its own, nascent strand. Inset shows a duplex scorpion that exchanges the stem–loop structure for a primer element terminally labelled with the fluorophore and a separate complementary oligonucleotide labelled with a quencher at the 5′ terminus.