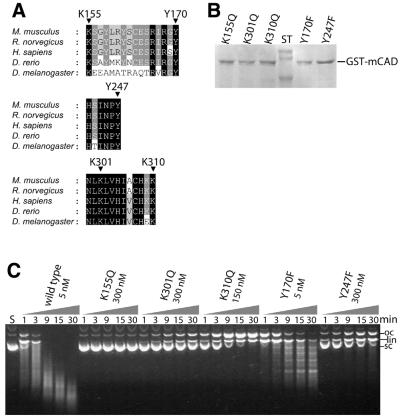
Figure 2.
Mutational analysis of variants with exchanged conserved lysine and tyrosine residues of GST-mCAD. (A) Amino acid residues Lys155, Lys301 and Lys310 as well as Tyr170 and Tyr247, which were exchanged to glutamine or phenylalanine, respectively, are fully conserved among the apoptotic nucleases known from the five indicated species [Mus musculus (GenBank accession nos AB009377, NM_007859), Rattus norvegicus (GenBank accession no. AF136598), Homo sapiens (GenBank accession nos AF064019, AF039210, AB013918, NM_004402), Danio rerio (GenBank accession no. AF286179) and Drosophila melanogaster (GenBank accession nos AF149797, AB036773)]. Shading is according to the Blosum 62 scoring matrix, with black shading for 100%, dark grey for 80% and light grey for 60% conserved amino acid residues. (B) SDS–PAGE analysis of variants of GST-mCAD that were produced as a complex with hICAD-L in E.coli and activated by treating the complex with recombinant caspase-3. (C) DNA cleavage activity of the GST-mCAD variants were measured by the disappearance of supercoiled plasmid DNA, analysed by agarose gel electrophoresis. GST-mCAD-Y170F is the only variant that retains wild-type activity with respect to the cleavage of supercoiled DNA, whereas all other variants exhibit strongly reduced cleavage activities. GST-mCAD-Y170F produces a pattern of fragments of defined length instead of randomly cut fragments appearing as a ‘smear’ (see wild-type GST-mCAD). Note the different concentrations of the variants as indicated.