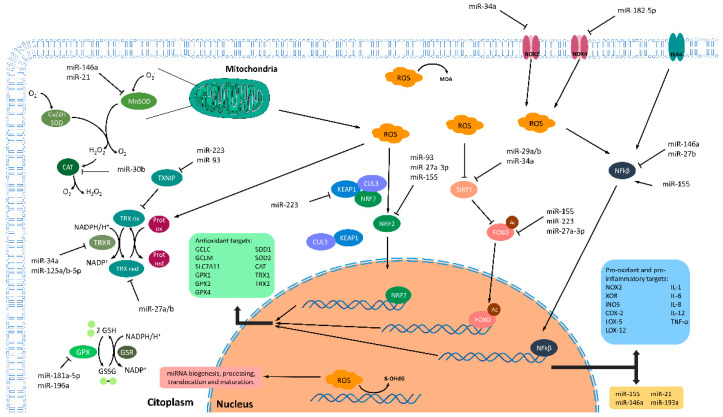
Figure 2.
Principal interconnections of ROS, miRNAs, and inflammation. Redox imbalance is achieved by the coordinated work of ROS production and antioxidant systems. In this regard, dysregulation of these synchronized roles can produce ROS accumulation and cellular damage. In the nucleus, ROS modulate multiple steps of miRNAs metabolism including biogenesis, processing, translocation, and maturation of these small RNAs. Defective or increased ROS levels alter the normal miRNAs metabolism, consequently altering many processes regulated by them. Furthermore, increased levels of ROS affect the composition of DNA by generation of an oxidized nucleoside 8-hydroxydeoxyguanosine (8-OHdG). Nuclear localization of the transcription factors nuclear factor kappa B (NF-KB), forkhead box protein O (FOXO), and nuclear factor erythroid 2-related factor 2 (NRF2) can modulate the transcription of different antioxidant targets (green box). Furthermore, NF-KB can also modulate the pro-oxidant and pro-inflammatory targets (blue box) and the expression of some inflamma-miRNAs (yellow box). The abundance of these transcription factors in the cytoplasm, where they stay as inactive forms, is regulated by some miRNAs such as the inflamma-miR miR-155 or the family of miR-27a/b. In addition, ROS and inflammatory signals promote the nuclear translocation of these transcription factors. ROS generation is a consequence of both normal mitochondrial metabolism and inflammatory process that should be eliminated by the action of antioxidants system. In this regard, different miRNAs were described either as oxidative stress-responsive miRNAs, as is the case of miR-34a-5p, miR-150-3p, miR-155, and miR-223, or as a regulators of antioxidant enzymes (such as thioredoxin family, catalase, superoxide dismutase, glutathione peroxidases family, and glutathione metabolism-related enzymes). Induction (arrows) or inhibition (blunt-end arrows). Glutathione peroxidase (GPX); Glutathione reductase (GSR); Reduced glutathione (GSH); Oxidized glutathione (GSSG); Reactive oxygen species (ROS); Cullin-3 (CUL3); Kelch-like ECH-associated protein 1 (KEAP1); NAD-dependent protein deacetylase sirtuin-1 (SIRT1); Toll-like receptor 4 (TLR4); NADPH oxidase 2 (NOX2); NADPH oxidase 4 (NOX4); Malondialdehyde (MDA); Thioredoxin 1 (TRX1) and 2 (TRX2); Thioredoxin reductase (TRXR); Thioredoxin interacting protein (TXNIP); Catalase (CAT); Cu/Zn superoxide dismutase (Cu/Zn SOD; SOD1); Mn superoxide dismutase (Mn SOD; SOD2); Glutamate-cysteine ligase regulatory subunit (GCLM); Glutamate-cysteine ligase catalytic subunit (GCLC); Cystine/glutamate transporter (SLCA11); Xanthine oxidoreductase (XOR); Nitric oxide synthase, inducible (iNOs); Cytochrome c oxidase subunit 2 (COX-2); Polyunsaturated fatty acid 5-lipoxygenase (LOX-5); Polyunsaturated fatty acid lipoxygenase (LOX-12); Interleukin-1 (IL-1); Interleukin-6 (IL-6); Interleukin- (IL-8); Interleukin-1 (IL-12); Tumor necrosis factor alpha (TNF-α).