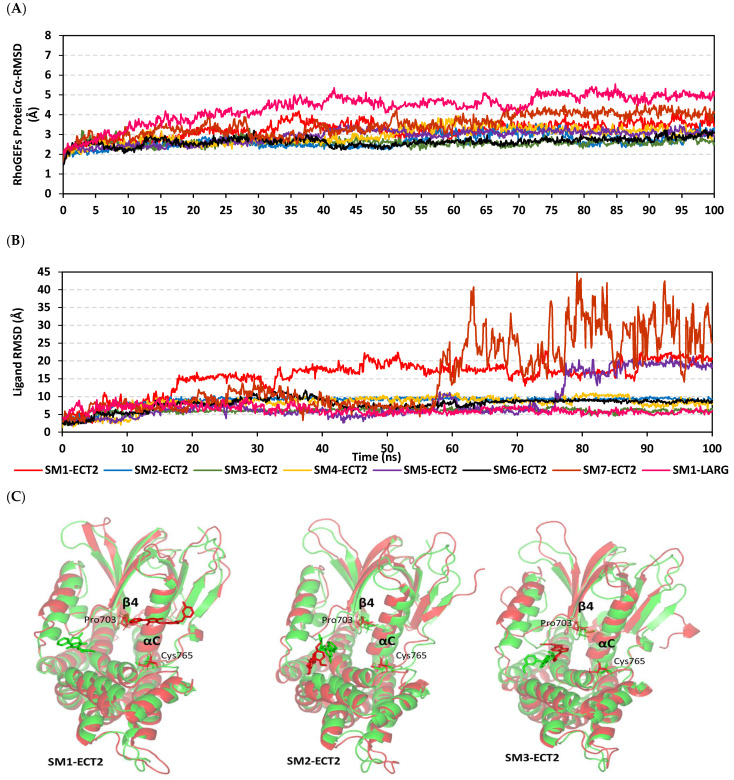
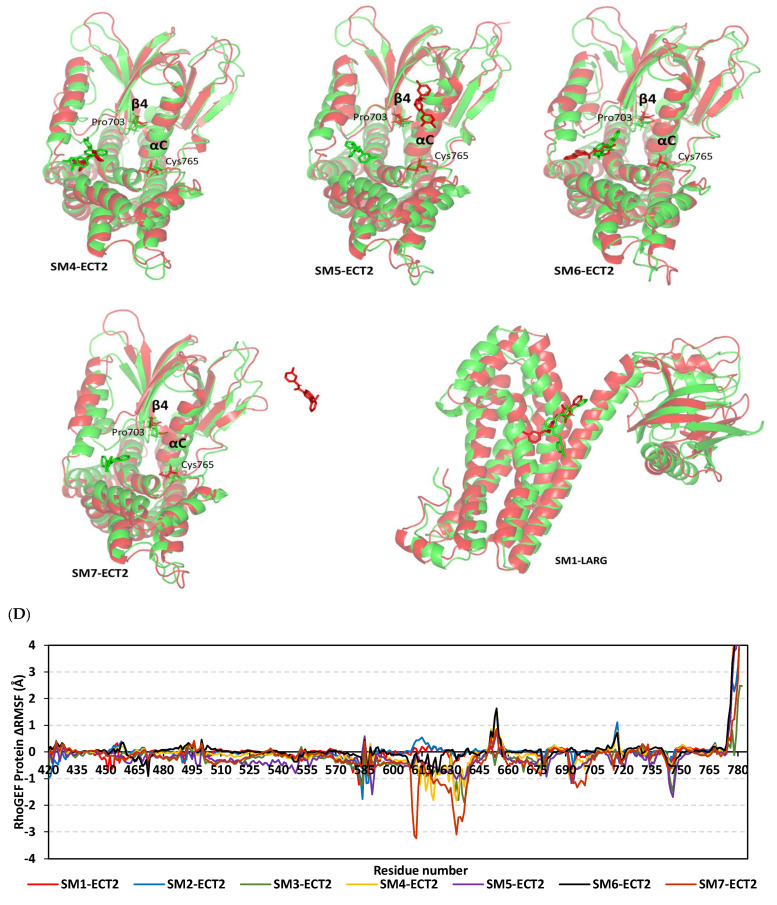
Figure 12.
Thermodynamic stability of the simulated small molecules bound to RhoGEFs. (A) Alpha-carbon atom RMSDs for RhoGEF proteins. (B) Sole ligand RMSDs in relation to simulation time frames in nanoseconds (ns). (C) Overlaid ligand/RhoGEF trajectories at initial and final time frames. Ligands (sticks) and bound RhoGEF proteins (cartoon) are colored green and red with respect to 0 ns and 100 ns extracted frames. For ECT2 proteins, Pro703 β4-sheet tip and C-terminal Cys765 at the PH domain are annotated. (D) Difference RMSF (ΔRMSF) trajectories for ECT2 target proteins along the whole molecular dynamic simulations in relation to apo/unliganded state.