Table 1.
Dissected docking–binding interactions for small molecules at DH-PH catalytic interface of RhoGEF ECT2 and/or LARGE.
| Small Molecules | Docking Binding Energy (Kcal.mol−1; RMSD Å) | H-Bond Interaction (Length Å/angle°) | Hydrophobic Interaction | π-Driven Interaction (Length Å) | van der Waal with Side Chain Carbons | |
|---|---|---|---|---|---|---|
| Code-Target | Chemical Structure | |||||
| SM1-ECT2 |
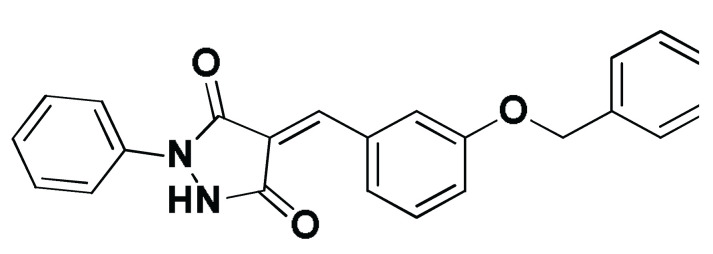
|
−5.824 (1.890) |
Lys527; 2.5/121 Gln567; 2.4/159 Asp610; 2.9/140 Arg612; 3.0/126 |
Val520, Phe523, Val559, Ile563, Val566, Ala616, His638 | Phe523; 5.1 Arg612; 3.2 |
Glu524 (Cβ,Cγ) Lys613 (Cγ,Cδ) |
| SM2-ECT2 |
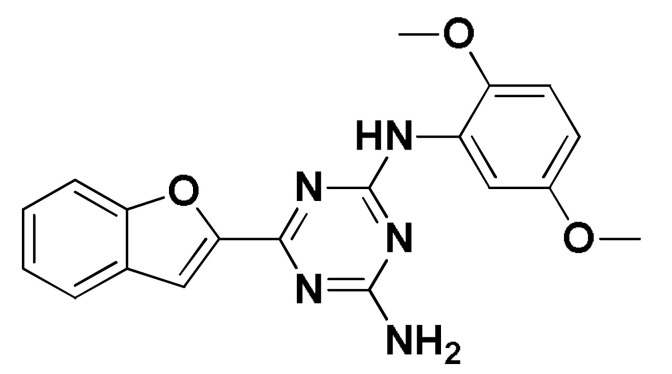
|
−6.369 (1.370) |
Glu524; 2.6/124 Lys527; 1.9/158 Lys527; 2.4/129 Arg612; 2.7/125 |
Val520, Phe523, Ile563, Val566, Pro570, Ile607, Ala616, His638 | Phe523; 5.0 Arg612; 3.1 Arg612; 2.0 |
Glu524 (Cβ,Cγ) Lys613 (Cβ,Cγ,Cδ) |
| SM3-ECT2 |
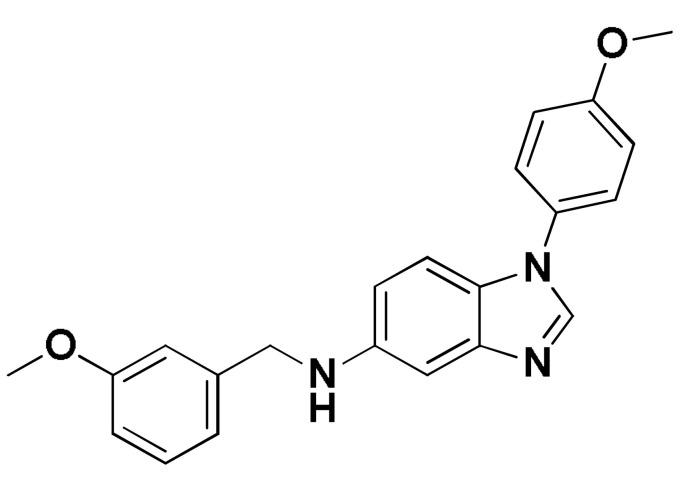
|
−6.061 (1.207) |
Gln567; 3.2/133 Arg612; 2.6/130 Arg612; 2.4/128 |
Val520, Phe523, Ile563, Val566, Pro570, Ile607, Ala616, His638 | Phe523; 4.9 Arg612; 3.1 |
Glu524 (Cβ,Cγ) Lys613 (Cγ,Cδ) |
| SM4-ECT2 |
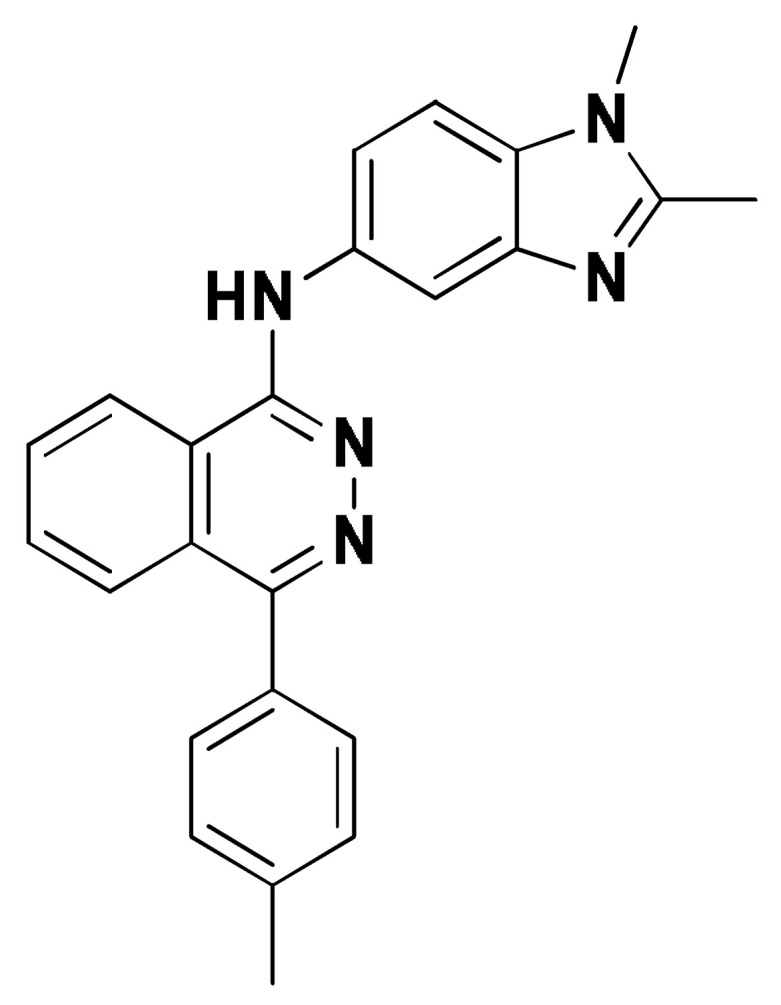
|
−5.855 (1.078) |
Gln567; 3.2/133 Arg612; 2.7/121 Arg612; 2.3/131 |
Phe523, Val559, Ile563, Val566, Ala616, His638 | Phe523; 5.0 Gln567; 5.1 Arg612; 3.1 Arg612; 3.0 |
Glu524 (Cβ,Cγ) Lys613 (Cβ,Cγ,Cδ) |
| SM5-ECT2 |
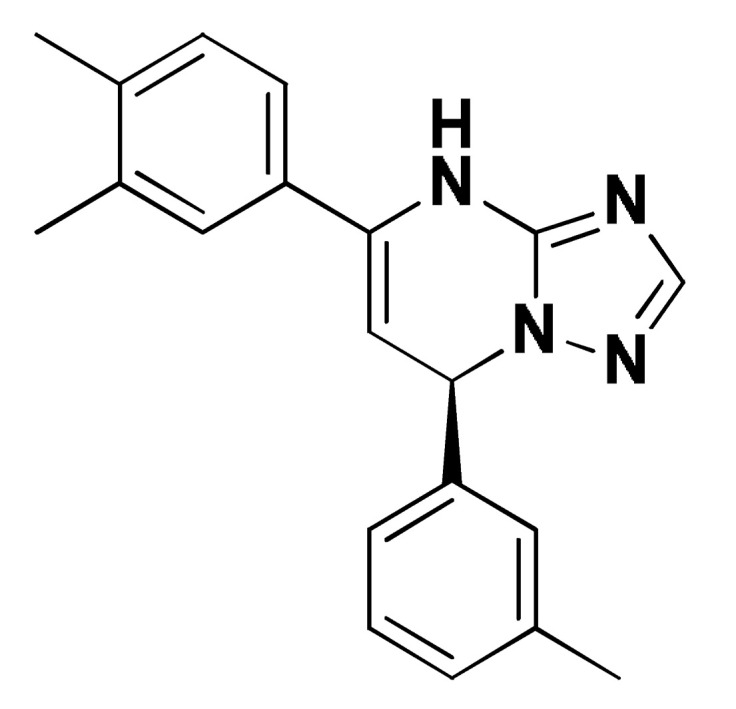
|
−4.972 (1.623) |
Arg612; 1.3/143 Arg612; 3.2/126 |
Val520, Phe523, Ile563, Val566, Pro570, Ile607, Ala616, His638 | Gln567; 3.7 Lys527; 3.0 Arg612; 2.7 |
Glu524 (Cγ) Lys613 (Cγ,Cδ) |
| SM6-ECT2 |
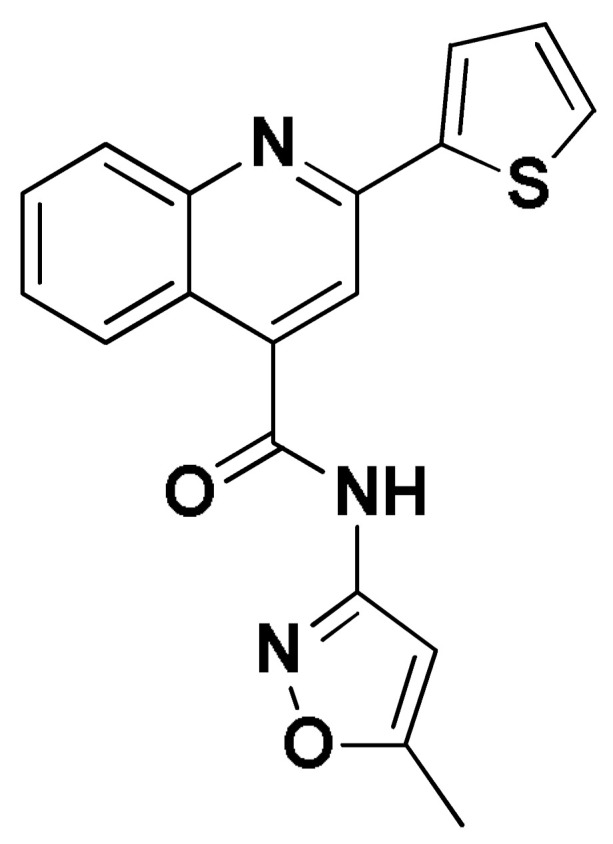
|
−4.853 (1.569) |
Arg612; 2.5/131 Arg612; 2.7/129 |
Phe523, Val559, Ile563, Ala616, His638, His759 | Phe523; 4.8 Glu567; 3.9 Arg612; 2.3 |
Glu524 (Cβ,Cγ) Lys613 (Cγ,Cδ) |
| SM7-ECT2 |
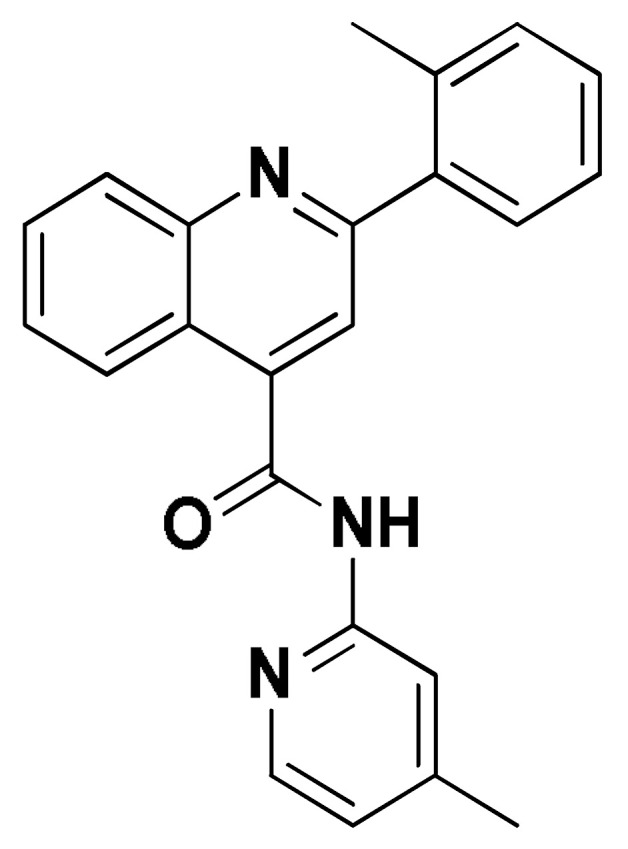
|
−5.469 (1.012) |
Gln567; 2.4/159 Asp610; 2.9/140 Arg612; 3.0/120 Lys527; 2.5/126 |
Val520, Phe523, Val559, Ile563, Val566, Ala616, His638 | Phe523; 4.8 Lys527; 4.7 Arg612; 3.2 |
Glu524 (Cβ,Cγ) Lys613 (Cβ,Cγ) |
| SM1-LARG |
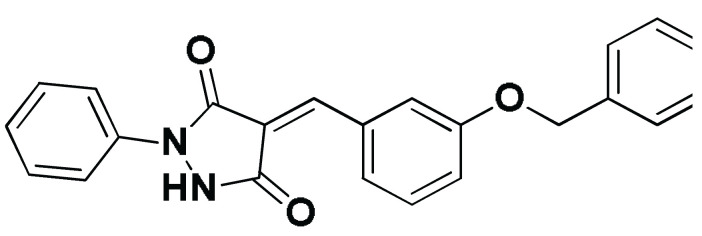
|
−5.534 (1.065) |
Pro892; 3.2/146 Arg986; 2.5/143 Arg986; 2.1/159 |
Cys888, Phe892, Leu895, Met934, Leu937, Leu971, Val974, Val978 | Phe893; 4.9 | Glu896 (Cβ,Cγ) Asn975 (Cβ) Gln985 (Cβ,Cγ) |
