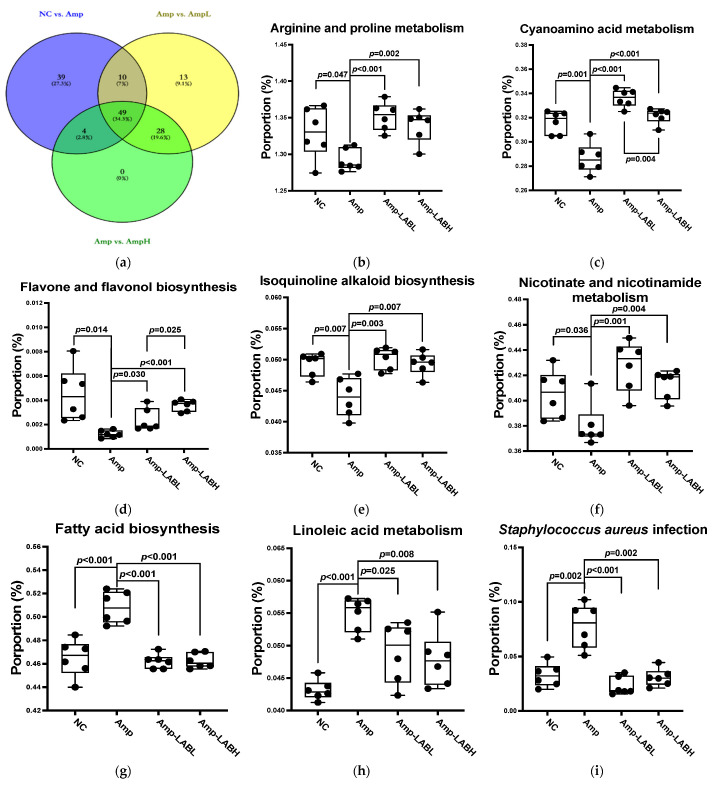
Figure 5.
Effect of Lactobacillus spp. on gut microbes that regulate functional pathways in C57BL/6J mice. Based on the analysis of 16S ribosomal RNA V3-V4 gene expression in the ileum, PICRUSt2 software was used for functional pathway prediction. (a) Venn diagram analysis of different functional pathways between the four groups. The dramatic difference between the Amp and LAB groups includes 49 pathways, such as (b) arginine and proline metabolism, (c) cyanoamino acid metabolism, (d) flavone and flavonol biosynthesis, (e) isoquinoline alkaloid biosynthesis, (f) nicotinate and nicotinamide metabolism, (g) fatty acid biosynthesis, (h) linoleic acid metabolism, and (i) Staphylococcus aureus infection.