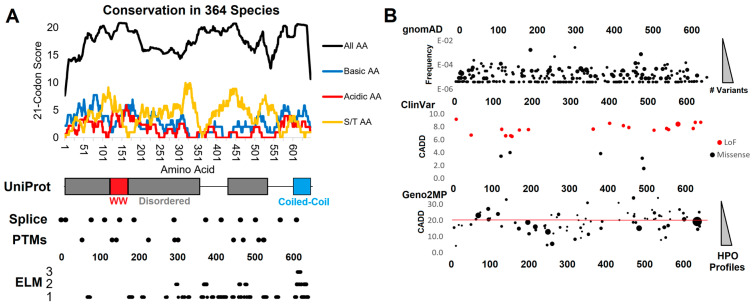
Figure 1.
Genomic analysis of WAC. (A) Compiled genomics insights for WAC. On top is the conservation of 364 species of WAC protein. The conservation score of all amino acids (black) is based on the amino acid % of species conserved at each amino acid (0—no conservation, 1–100% conserved), placed on a 21 codon sliding window such that each site is added to 10 upstream and 10 downstream. The basic (blue, R/K/H), acidic (red, D/E), or S/T (orange) amino acid scores are calculated using only these amino acids for the same 21 codon window. Below is conservation of the amino acid locations (x-axis) for UniProt annotated domains/motifs, splice sites, posttranslational modifications (PTMs), and ELM annotated motifs (y-axis lists the number of annotations at each amino acid). (B) Extracted genomic variants from gnomAD (y-axis list allele frequency, bubble size is the number of unique variants), ClinVar (y-axis is the CADD functional score, color represents the type of variant), and Geno2MP variants (y-axis is the CADD score, bubble size the number of human phenotype profiles). Abbreviations—AA, amino acid; R, arginine; S, serine; T, threonine; LoF, loss-of-function; ELM, eukaryotic linear motif; HPO, human phenotype ontology.