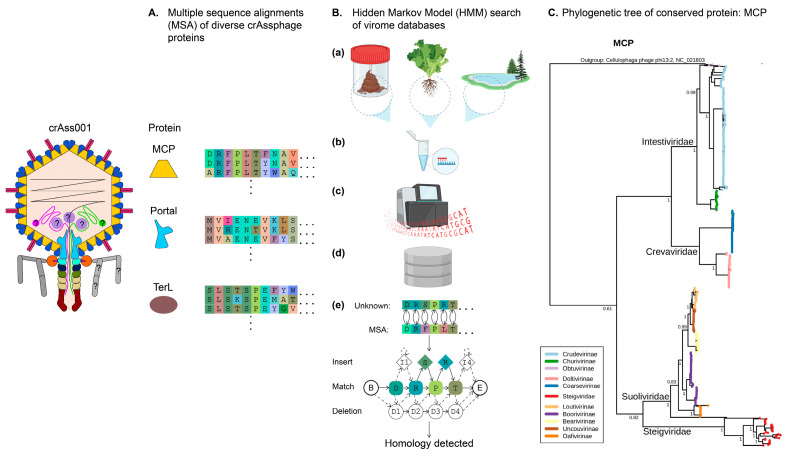
Figure 1.
Bioinformatic approaches to discovering and classifying crAss-like phages. (A), Multiple sequence alignments (MSA) of the evolutionary conserved phage proteins such as major capsid protein (MCP), portal protein, and terminase large subunit (TerL) contain signatures specific to the order Crassvirales. MSAs are converted to profile HMMs (Hidden Markov Models). (B), Discovery and classification of new Crassvirales. Candidate genomes are collected from varied environments such as stool, soil, or aquatic biomes (a); shotgun sequencing and assembly of total or VLP-enriched DNA (b,c) yields large assembly databases (d); translated proteins are compared to MSAs of conserved Crassvirales proteins through sensitive position-specific homology detection algorithms (PSI-BLAST, HMM, and HHM). (C), Using MCP as a taxonomic marker protein, a maximum likelihood phylogenetic tree can be constructed to infer relatedness between families (main first-order branch names) of crAss-like phages [48].