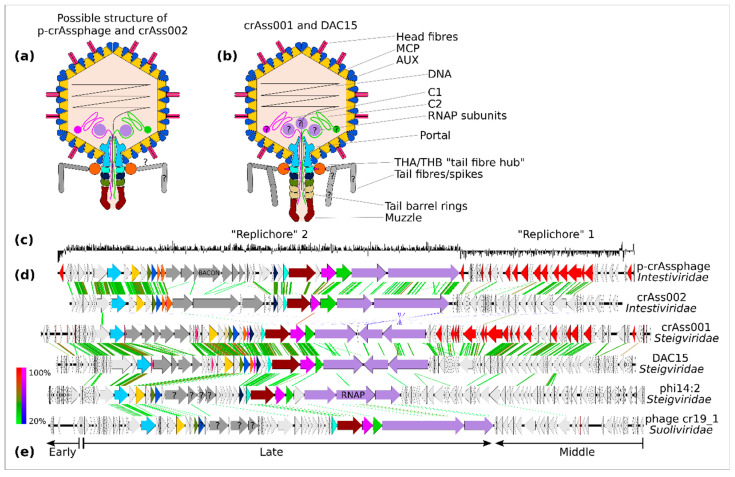
Figure 2.
Virion and genome structure of the typical representatives of the order Crassvirales. A schematic structure of Intestiviridae virions (a) is extrapolated from the cryoEM structural data available for Steigviridae (b). Colours of the protein subunits (a,b) are consistent with ORF colouring (d). GC-skew graph of the p-crAssphage genomes shows clear separation into two possible replichores (c). Despite significant divergence in the amino acid sequence of the encoded proteins, all families show clear genome collinearity (d). Three transcriptional modules can be identified: early (regulatory), middle (DNA replication), and late (structural proteins and other virion components), as exemplified by a Suoliviridae genome (e).