Abstract
Danio rerio (zebrafish), traditionally used in forward genetic screens, has in the last decade become a popular model for reverse genetic studies with the introduction of TALENS, zinc finger nucleases, and CRISPR/Cas9. Unexpectedly, homozygous frameshift mutations generated by these tools frequently result in phenotypes that are less penetrant than those seen in embryos injected with antisense morpholino oligonucleotides targeting the same gene. One explanation for the difference is that some frameshift mutations result in nonsense-mediated decay of the gene transcript, a process which can induce expression of homologous genes. This form of genetic compensation, called transcriptional adaptation, does not occur when the mutant allele results in no RNA transcripts being produced from the targeted gene. Such RNA-less mutants can be generated by deleting a gene’s promoter using a pair of guide RNAs and Cas9 protein. Here, we present a protocol and use it to generate alleles of arhgap29b and slc41a1 that lack detectable zygotic transcription. In the case of the arhgap29b mutant, an emerging phenotype did not segregate with the promoter deletion mutation, highlighting the potential for off-target mutagenesis with these tools. In summary, this chapter describes a method to generate zebrafish mutants that avoid a form of genetic compensation that occurs in many frameshift mutants.
Keywords: CRISPR/Cas9, Zebrafish, arhgap29b, RNA-less mutant
1. Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR) guide RNAs combined with CRISPR-associated 9 (Cas9) protein is a robust tool for genetic engineering in cell lines and animal model systems, and its application in gene therapy is already underway for several diseases [1–3]. The CRISPR/Cas9 system can introduce site-specific double-strand breaks, which are then repaired through nonhomologous end joining (NHEJ), homologous recombination, or by a potentially distinct mechanism called homology-directed repair. As in other model systems, in zebrafish NHEJ is robust, but leads to small insertions or deletions (indels) in DNA sequence. CRISPR/Cas9-mediated mutagenesis in zebrafish was first applied to generate indels in exons, leading to frameshifts and premature stop codons [4–8]. Additionally, pairs of guide RNAs (gRNAs), which are used to direct the Cas9 endonuclease to specific locations in the genome, were used to generate larger deletions, 7–50 kilobases long, that were targeted albeit with imprecise edges [9, 10]. Later homologous recombination and homology-directed repair, which are distinguished functionally by the length of the homology arms in the template used to invoke them, were deployed to generate more accurate editing of genomic DNA [11–13]. Two improvements in the use of CRISPR/Cas9-mediated gene editing in zebrafish were, first, the discovery that delivery of Cas9 in protein form, rather than as mRNA, facilitated more rapid double-strand breaks [14, 15]. Second, it was found that chemically modifying the gene-targeting CRISPR RNA (crRNA) to protect it from cellular RNases increased the efficiency of generating double-strand breaks [16, 17].
The emergence of gene-targeting technologies led to multiple laboratories making the same puzzling observation; the phenotype of a targeted mutant was often less severe than that in embryos injected with antisense morpholino oligonucleotides (MO) targeting the identical gene [18]. In some instances, this phenomenon probably reflects the MO disrupting expression of an unintended gene (i.e., off-target effect) [19]. In other instances, it reflects increased expression of genes homologous to the targeted one that occurs in frameshift mutants, but not in the corresponding “morphants” [20]. The elevated expression of the homologs depends on the incomplete gene transcripts that result from nonsense-mediated decay; the underlying mechanism, which remains poorly understood, is called transcriptional adaptation [20]. A means to avoid transcriptional adaptation, and consequently to reveal the function of the targeted genes, is to generate loss-of-function alleles that yield no transcript, i.e., RNA-less alleles [20]. This chapter describes the use of CRISPR/Cas9 to make RNA-less alleles in zebrafish. We targeted the arhgap29b and slc41a1 genes. The former is of interest to our group because of the association of its ortholog to orofacial cleft [21–23], and the other because it encodes a magnesium transporter that may be important in differentiation of dopaminergic neurons [24]. We used pairs of gRNAs to generate promoter-less mutations of each gene (separately) and showed that levels of the corresponding mRNA in homozygous mutants are, in one case (slc41a1), undetectable, and in the other (arhgap29b), are very low and we presume reflect residual maternal deposits. Interestingly, a phenotype emerged in the progeny of fish heterozygous for the arhgap29b promoter-deletion that did not segregate with this mutation. This suggests the CRISPR/Cas9 reagents yielded an off-target mutation in addition to the intended one. Collectively, promoter-deletion is an approach in using CRISPR/Cas9 gene editing to generate null alleles in zebrafish, which overcomes the challenge of transcriptional adaptation that results from earlier methods of employing this technology. Nonetheless the potential for off-target effects of CRISPR/Cas9 technology remains a challenge to the field.
2. Materials
Alt-R® CRISPR-Cas9 tracrRNA.
Gene-specific crRNA; IDT (sequences are provided in Table 1).
IDTE (1× TE Solution pH 7.5); IDT.
Duplex buffer; catalog # 1072570; IDT.
SP Cas9 protein.
DNA lysis buffer: 10 mM Tris–HCl, pH 8, 1 mM EDTA—pH 8, 0.01% SDS, 100 mM NaCl, and 0.1 mg/ml Proteinase K (see Note 1). Store at −20° C.
PCR screening primers (sequences are provided in Table 2 and see Note 2).
PCR reagents (PCR buffer, dNTPs, Taq polymerase).
1% Tricaine: Dissolve 0.5 g of tricaine in 50 ml of fish water (see Note 3). Store at room temperature (RT).
PBST: 1× PBS with 0.1% Tween-20.
Trizol reagent.
Turbo DNA-free kit.
High-capacity cDNA reverse transcription kit.
Gene-specific RT-PCR primers (sequences are provided in Table 3 and see Note 2).
iTaq Universal SYBR Green Supermix.
Table 1.
crRNA sequences and their scores
| Gene | Guide | Sequence | PAM | On-target score | Off-target score |
|---|---|---|---|---|---|
| arhgap29b | Left guide | ATTTCAAGCTTTCAAGTACT | GGG | 84 | 75 |
| Right guide | TAACGTGACTGTTTCAATCG | TGG | 71 | 91 | |
| slc41a1 | Left guide | TTCTTGGACTACTACGAGAA | AGG | 59 | 93 |
| Right guide | GAGAAGGACGTTTCCTTAAG | TGG | 65 | 92 |
Table 2.
Primers used for screening
| Primers used for screening | Sequences | Amplicon length | |
|---|---|---|---|
| arhgap29b | Forward primer | GCAATATAATGTCCTTCGGCTAA | WT-420 bp, Het-420 bp and 368 bp, Mut-368 bp |
| Reverse primer | GGTTACGTGACTGCCTTTGTC | ||
| Internal forward primer | GCTAGCAGCGCTAAAGTTCAT | ||
| slc41a1 | Forward primer | CAGCTTTCGGAAGCACTTAGG | WT-498 bp, Het-498 bp and 198 bp, Mut-198 bp |
| Reverse primer | CCAGCAGGAAAGGGATCAAAAC | ||
| Internal reverse primer | GGCTGTCAACTCCGATCAGT | ||
Table 3.
Primers used for qPCR
| Primers for qPCR | Forward qPCR primer sequence | Reverse qPCR primer sequence |
|---|---|---|
| arhgap29b | CATGTACTGCTCCAAACGGC | CTCCGGGCCTTCTCGTATTC |
| slc41a1 | AGCCAGTCGTATCTCCACCT | TTCACACCAGACCCAAAGAA |
| beta actin | CGAGCAGGAGATGGGAACC | CAACGGAAACGCTCATTGC |
3. Methods
3.1. Establish the Promoter(s) Sequence of the Gene of Interest
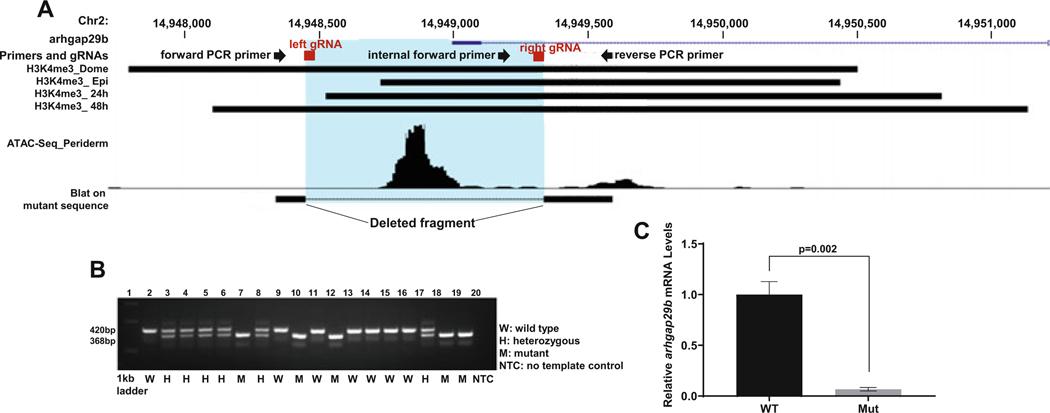
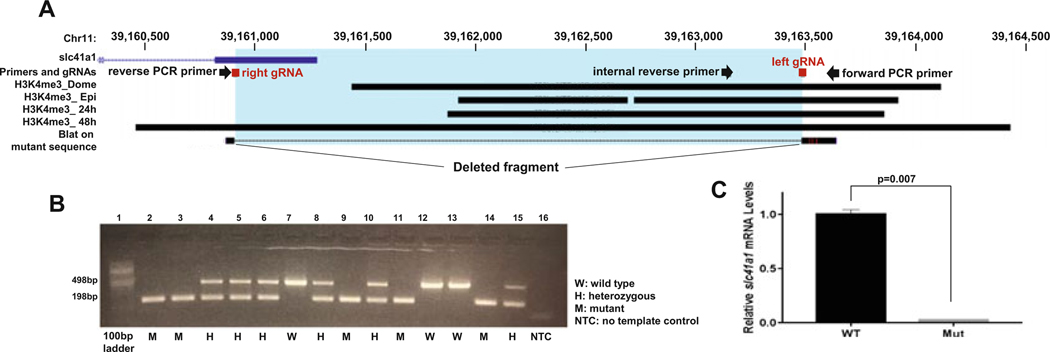
Identify the promoter of the gene of interest by using the corresponding trimethylation at histone H3, lysine 4 (H3K4me3), and/or Assay for Transposase-Accessible Chromatin sequencing (ATAC-seq) data and load them on the UCSC genome browser (see Notes 4, 5, 6, and 7) (Figs. 1a and 2a).
Having identified the promoter region, look for the guide RNAs (gRNAs) positioned within 1 kb sequences on either side of the H3K4me3 and/or ATAC-seq peak (see Note 8).
Fig. 1.
RNA-less allele of arhgap29b in zebrafish embryos. (a) H3K4me3 data from whole embryos, and ATAC seq data, from isolated periderm, around the presumed transcription start site of arhgap29b (inferred from the Refseq gene model). There is broad H3K4me3 signal over the transcription start site; by contrast, the ATAC seq signal is much narrower. These marks were used to determine where to target the guide RNAs (gRNAs). gRNAs and primer sequences used for screening are marked with a block and arrow, respectively. The blue region highlights the deleted fragment after CRISPR/Cas9 targeting. (b) Agarose gel electrophoresis of zebrafish embryos from F2 generated by crossing F1 fish in which the arhgap29b promoter was targeted. Gel shows a wild-type (420 bp) and mutant-specific (368 bp) bands. The mutant band was sequenced, and a BLAT search on the UCSC browser showed clear deletion of the promoter region due to arhgap29b-specific gRNAs, highlighted in blue in panel a. (c) Bar graph of relative levels of arhgap29b mRNA in a pool of WT and a pool of mutant embryos from F2, generated by CRISPR/Cas9 targeting as described in a and analyzed by qPCR. Expression levels of arhgap29b are normalized to expression of beta-actin. Bars represent the mean and standard deviation of three independent experiments. P value determined by student’s t-test
Fig. 2.
RNA-less allele of slc41a1 in zebrafish embryos. (a) H3K4me3 data from whole embryos at the presumed transcription start site of slc41a1 (inferred from the refseq gene model). Guide RNAs (gRNAs) and primers sequences used for screening are marked with a block and arrow, respectively. The blue region highlights the deleted fragment after CRISPR/Cas9 targeting. (b) Agarose gel electrophoresis of zebrafish embryos from F2 generated by crossing F1 fish in which the slc41a1 promoter was targeted. Gel shows a wild-type (498 bp) and mutant-specific (198 bp) bands. The mutant band was sequenced, and a BLAT search on the UCSC browser showed clear deletion of the promoter region due to slc41a1-specific gRNAs, highlighted in blue in panel a. (c) Bar graph of relative levels of slc41a1 mRNA in a pool of wild-type and a pool of mutant embryos, generated by CRISPR/Cas9 targeting as described in a and analysed by qRT-PCR. Expression levels of slc41a1 are normalized to expression of beta-actin. Bars represent the mean and standard deviation of three independent experiments. P value determined by student’s t-test
Once these sequences are identified, they can be uploaded to a gRNA design webtool of your choice (see next section).
3.2. Design gRNAs (See Note 9)
For the sequence upstream of the promoter, which we will refer to as the “left crRNA,” download 1 kb of target sequence from the region to the left of the promoter locus (i.e., defined by the region of open chromatin or island of H3K4me3 signal) to be deleted. (We use “left” and “right” here, rather than upstream and downstream, because the promoter may be pointed in either direction.) This sequence should be in FASTA format and can be obtained from the UCSC genome browser.
Design gRNAs with an appropriate gRNA design webtool. We used the “Custom Alt-R® CRISPR-Cas9 guide RNA” design tool available on the IDT website and selected “Danio rerio” as the species from the dropdown menu. (https://www.idtdna.com/site/order/designtool/index/CRISPR_CUSTOM).
Enter the sequence or upload it as a text file. We entered the left-hand target sequence in FASTA format.
Select a desired crRNA sequence among the several suggested by the algorithm. We selected a crRNA sequence with high on-target and high off-target scores. In the IDT gRNA design tool platform, the higher off-target scores have a lower risk of targeting nonspecific sequences (see Note 10). Ideally this will be located outside of the H3K4me3 signal but in the two cases presented here the crRNAs were not completely outside of the H3K4me3 signal at all developmental stages assessed.
Repeat this process using 1 kb of target sequence to the right of the promoter to identify the sequence for the “right crRNA.”
Use a PCR primer design tool (e.g., Primer3) to design PCR primers that flank the desired sequence to be removed. We place the primer at least 50 bp from the gRNAs to assure the primer site is not deleted during nonhomologous end joining.
3.3. Prepare sgRNAs and RNP Complexes for Injection into Embryos
Gene-specific crRNAs and tracrRNA from IDT are available in different quantities. We ordered two nanomoles of each crRNA and five nanomoles of the generic tracrRNA. Starting with these amounts, we resuspended crRNAs and tracrRNAs in 20 μl and 50 μl of IDTE buffer, respectively (1× TE buffer, pH 7.5), which yielded a final concentration of 100 μM in both cases. These were stored at −80° C (see Note 11).
Generate separate solutions for the left and right crRNAs, each consisting of tracrRNA and the specific crRNA at a final concentration of 10 μM each. Pipette 2.5 μl of 100 μm tracrRNA and 2.5 μl of 100 μM crRNA into 20 μl of IDT duplex buffer.Vortex gently and briefly centrifuge the solutions.
Generate a single gRNA (sgRNA) for each crRNA by annealing the tracrRNA and crRNA; heat the solution from step 2 at 95 °C for 5 min and gradually allow it to cool to RT on the benchtop. This can be stored at −20 °C for 6 months and −80 °C for 1 year (see Note 12).
SpCas9 protein was supplied at 61 μM concentration. Pipette 1 μl of this stock into 3 μl of Cas9 dilution buffer for a final concentration of 15 μM. Make several 1 μl aliquots of 15 μM Cas9 and store at −80 °C.
3.4. Inject Embryos with gRNA
The night before the injection, set up zebrafish crosses by placing 2–3 wild-type males and 2–3 wild-type females in a tank separated by a divider (see Note 13).
On the morning of the injection, pull out the divider and wait for the eggs. It may take 30–40 min before eggs are laid. Pipette 1 μl of the “left” sgRNA, 1 μl of the “right” sgRNA, 1 μl of 15 μM Cas9 protein, 1 μl of phenol red into a micro-centrifuge tube, and 1 μl of nuclease-free water to final volume of 5 μl, and store on ice (see Note 14). Pipette to mix. This yields a final concentration of 2 μM for each sgRNA and 3 μM for Cas9 protein.
Incubate at RT for 5–10 min to allow ribonucleoprotein complexes to form.
Inject 5 nl of RNP complex to each embryo at the 1–4 cell stage. Inject at least 100 embryos; leave at least 10–20 embryos uninjected as controls.
Incubate injected and uninjected embryos in fish water at 28 °C incubator (see Note 15).
3.5. Genotype F0 Embryos
24 h after injection, randomly select 10 injected embryos and 4–5 uninjected embryos, dechorionate them, and place them individually in PCR tubes (we use strips of 8 tubes). Continue to incubate remaining embryos; if mutagenesis was successful, these animals can be used for breeding.
Perform a crude preparation of genomic DNA by adding 20 μl of DNA lysis buffer to each embryo in a PCR tube. Place the tubes in a thermal cycler and incubate at 55 °C for 50 min followed by 95 °C for 10 min. Store the DNA at 4 °C.
Genotype embryos by PCR with 1 μl of genomic DNA using PCR primers that flank the intended deletion (Table 2). Run the program on a thermal cycler with the following steps: 1 cycle: 95 °C for 5 min; 35 cycles: 95 °C for 20 s, 55 °C for 30 s, 72 °C for 30 s; 1 cycle: 72 °C for 5 min. Run the PCR product on a 2.5% agarose gel and look for the mutant-specific band size. Excise the mutant-specific band at the predicted size and confirm by sequencing (see Notes 16 and 17) (Figs. 1a, b and 2a, b).
Check the efficiency of the CRISPR/Cas9-mediated deletion with a three-primer PCR (see Notes 16 and 17) (Table 2, Figs. 1a, b, and 2a, b).
3.6. Generate and Raise F1 Fish
After confirming the intended promoter deletion is present in F0 embryos, raise the remaining F0 animals to breeding age. The efficiency of mutagenesis in the embryos can be used to predict how many F0 adults will be needed to achieve germline transmission of the mutation (see Note 18).
If desired, one can confirm the mutation is present in somatic tissue of F0 fish by clipping a fin at 1 month or older, harvesting genomic DNA from the fin, and performing PCR on the DNA with genotyping primers (see Note 19). For fin clipping, anesthetize the fish in a petri dish using anesthetizing solution (0.01% tricaine in fish water, see Note 20). Once, anesthetized (may take 20–60 s), use a spoon to place fish on parafilm and excise the caudal fin lobe. Lyse with the same protocol mentioned in Subheading 3.5, step 2. Once clipped, place the fish back into its tank with fish water (see Note 20).
To identify F0 fish that transmit mutations, cross individual mutant F0 fish to wild-type fish and screen pools of F1 embryos for the promoter mutation. Make 5 pools of 5 randomly selected F1 embryos at 24 hours post fertilization (hpf) each, make a crude genomic DNA preparation from each pool, and screen the DNA by PCR for the deletion. Confirm the presence of the mutation by sequencing the PCR band. If the mutation is detected, raise the remaining F1 embryos to adulthood.
At 1–2 months of age, genotype the F1 animals by fin clip and PCR and continue to raise those harbouring the mutation.
3.7. Generate F2 Embryos and Assess if Those with Abnormal Phenotypes Harbour the Targeted Promoter Mutation
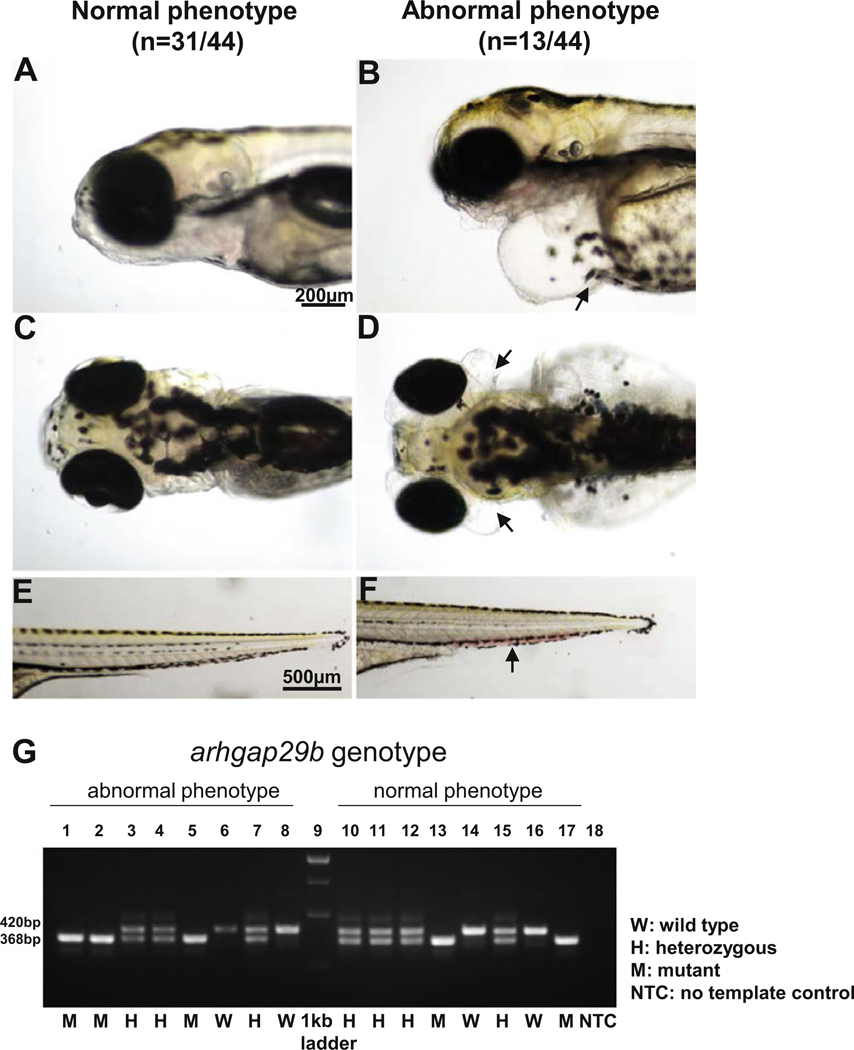
Cross F1 carriers then observe and record phenotypes in F2 embryos, looking for a consistent phenotype in approximately one-fourth of the clutch (Fig. 3) (see Note 21).
Score the genotype of individual F2 embryos in each phenotypic class (see Notes 22 and 23) (Fig. 3).
Fig. 3.
Phenotypes of arhgap29b mutant embryos. Mutant embryos from the in-cross of F1 carriers of the arhgap29b allele in which the promoter has been deleted show different phenotypes. In contrast to (a, c, e) sibling embryos with a normal phenotype, (b, d, f) those with a phenotypically abnormal phenotype show (b) severe edema near the heart, (d) a bubble below the eye, and (f) poor blood circulation. Scale bar in a is 200 μm which corresponds to panels b–d; scale bar in e is 500 μm, which corresponds to panel f. (g) Homozygous mutant genotypes were recorded from both groups (embryos with normal phenotype and abnormal phenotype) suggesting the abnormal phenotype is an off-target effect
3.8. Determine mRNA Levels of the Targeted Gene
Cross 2 fish from the F1 (or later) generation that are heterozygous carriers of the intended mutation. Raise F2 embryos to 3 days postfertilization (or another stage, depending when the expression level of the targeted gene is known to be high). Cut the embryos in half and place head and tail portions into individual PCR tubes with proper labeling. For DNA isolation, add 20 μl of DNA lysis solution to the tubes containing tails and for RNA isolation, add 30–50 μl of Trizol reagent to the tubes containing heads (or the reverse, depending where the transcript is expressed at highest levels). Store tubes containing heads at −80 °C.
Perform a genotyping screen using the DNA preparation from tail samples with the appropriate primers for each sample, as described in Subheading 3.5, steps 2 and 3.
Based on the results of the genotyping screen, pool a minimum of 20–30 heads with the homozygous mutant genotype into a single 1.5 ml tube for RNA isolation. Homogenize the heads and bring the final volume up to 1 ml with Trizol reagent. Process the same number of wild-type embryos as a control.
- Perform RNA isolation according to the manufacturer’s standard protocol.
- Briefly, incubate the homogenized embryo heads at RT for 5 min. Add 200 μl of chloroform/per 1 ml of Trizol reagent and vortex vigorously for 15–20 s. Allow samples to stand at RT for 10–15 min.
- Centrifuge the resulting mixture at 13,500 × g for 15 min at 4 °C. Centrifugation separates the mixture into three phases: a red organic phase (containing protein), an interphase (containing DNA), and a colorless upper aqueous phase (containing RNA).
- Carefully transfer the aqueous phase to a new tube and add an equal volume of isopropanol (500 μl); mix the tubes by inverting them up and down. Allow samples to stand at RT for 10 min, then centrifuge at 13,500 × g for 10 min at 4 °C (see Note 24).
- Discard the supernatant carefully and wash the RNA pellet by adding 1 ml of 75% ethanol followed by centrifugation at 13,500 × g for 5 min at 4 °C.
- Remove the supernatant, air-dry the pellet, and dissolve in 20 μl of nuclease-free water. Measure the quantity of RNA on a spectrophotometer (e.g., Nanodrop 2000, Thermo-Fisher) and store at −80 °C, or proceed immediately to the next step.
Perform DNase treatment of RNA samples from step 4e. Add 1 μl of DNase1 for a maximum of 10 μg RNA, 3 μl of Turbo DNase buffer, and bring the final volume to 30 μl with nuclease-free water. Incubate the tubes at 37 °C for 30 min. Add 3 μl of inactivation reagent provided in the kit, mix well by pipetting, and incubate at RT for 5 min (see Note 25). Flick the tubes occasionally for intermittent mixing. Centrifuge the tubes at 13,500 × g for 90 s at RT. Transfer 20–25 μl of supernatant to a fresh 1.5 ml tube (see Note 26). Measure the quantity of DNase-treated RNA. Store at −80 °C.
Carry out first-strand cDNA synthesis from RNA samples using a high-capacity cDNA reverse transcription kit as per the manufacturer’s protocol. Use 1 μg of RNA for cDNA synthesis in a total reaction volume of 20 μl. Store the cDNA samples at −20 °C.
Perform qPCR using SybrGreen qPCR master mix. For a single reaction, use 5 μl of 2× SybrGreen, 0.2 μl of gene-specific forward and reverse primers (2 μM each, Table 3), and 1 μl of cDNA in a total reaction volume of 10 μl. Each reaction should be performed in triplicate. Prepare the master mix accordingly.
Simultaneously set up the reaction for housekeeping genes (e.g., actb1). Run the final reaction in a real-time PCR machine as per the manufacturer’s protocol.
Analyze the data using a suitable quantitation algorithm for real-time PCR (see Note 27). Calculate mean values with their standard deviations with respect to replicate experiments, and analyze for statistical significance of differences using a t-test (see Note 28).
4 Notes
Mix 10 μl of 1 M Tris–HCl, pH 8, 2 μl of 0.5 M EDTA, 1 μl of 10% SDS, 20 μl of 5 M NaCl, 10 μl of 10 mg/ml Proteinase K, and 957 μl of water. Use ultrapure water (prepared by purifying deionized water to attain a sensitivity of 18 MΩ-cm at 25 °C) throughout the protocol.
Resuspend the primers in a desired volume of water at 100 μM concentration (stock concentration) and dilute it further at a 1:10 ratio in water to get a 10 μM working concentration. Store at −20 °C.
Prepare fish water by dissolving instant ocean sea salt at a concentration of 0.5 g/L in distilled water.
Trimethylation at histone H3, lysine 4 (H3K4me3) is a characteristic of promoter regions [25], and ChIP-seq analysis of H3K4me3 has been reported in several stages of zebrafish embryos [26]. Load these or more up-to-date data into the UCSC genome browser as a custom track, i.e., load NCBI Gene Expression Omnibus sample numbers GSM915189 (H3K4me3_dome stage), GSM915190 (H3K4me3_80% epiboly stage), GSM915191 (H3K4me3_24 hpf), and GSM915192 (H3K4me3_48hpf) as custom tracks at UCSC (Figs. 1A and 2A).
ChIP-seq analysis of H3K4me3 at the arhgap29b locus shows a strong peak in zebrafish at the dome stage, 80% epiboly stage, 24 hpf and 48 hpf. Given that the H3K4me3 mark extends over a broad chromatin region, we also examined the arhgap29b promoter region for evidence of open chromatin by analyzing Assay for Transposase-Accessible Chromatin sequencing (ATAC-seq) data and DNase I hypersensitive sites, available through the at NCBI Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/).
The arhgap29b gene is expressed in the enveloping layer, so we specifically assessed ATAC-seq data that was obtained from isolated periderm [27]. An ATAC-seq peak was detected in isolated periderm at the transcriptional start site of arhgap29b (DanRef7, chr2: 14948752–14949078), and this was our target to delete.
We reasoned that deleting the region of open chromatin, where transcription factors are most likely to bind, was sufficient to eliminate promoter activity; however, some transcription factors bind DNA within nucleosomes, so it may be a preferred general practice to delete the entire island of H3K4me3 signal.
It is important to note that many genes utilize more than one promoter [28, 29]. H3K4me3 data from whole embryos should reveal the predominant promoter deployed at the stage the data were collected. However, given the possibility that a minor promoter becomes deployed after a major one is deleted, it is essential to confirm that there is no detectable mRNA in homozygous mutants.
gRNAs consist of a complex of two RNAs that are annealed together: one is a generic transactivating CRISPR RNA (tracrRNA) that recruits the Cas9 protein and the other is a CRISPR RNA (crRNA) that targets a specific sequence of interest [30].
IDT shows a square box with several predicted gRNA designs. Sequences that lie in the white portion of the box in the upper right corner are the best to choose. All sequences in red tend to have a higher risk of nonspecific targets and are not considered good gRNAs.
Before resuspending, briefly spin down the tubes with tracrRNA and crRNA. Incubating the resuspended crRNA and tracrRNA at 50 °C for 10 min helps to dissolve the RNA properly.
Each crRNA should be separately annealed with tracrRNA.
When making a mutant strain, it is best practice to start with an inbred strain. We favor the NHGRI-1 line because it has been sequenced, which makes it trivial to design PCR primers and gRNAs that are a perfect match to the genome [31]. The two examples presented here were generated in outbred fish.
Always thaw the sgRNAs, crRNAs, tracrRNAs, and Cas9 protein on ice before proceeding with any reaction set-up.
Change the fish water every 24 h for 2–3 days.
For genotyping a deletion allele by PCR, the simplest approach is to use a pair of primers flanking the deletion. However, in our experience, PCR on genomic DNA from heterozygous mutants often yields only the band from the deleted chromosome because smaller PCR products amplify much more efficiently than larger ones. Therefore, we use a three-primer design with two primers flanking the deletion and one within it, so bands of distinct but comparable sizes (200–500 bp) are amplified from intact chromosomes and those with the deletion.
In the case of the arhgap29b gene, our forward and reverse primers yielded a 1253 bp band from wild-type genomic DNA, and a 368 bp band from genomic DNA that had been successfully cut by both gRNAs (Table 2). Using an internal forward primer in addition to the two flanking primers, and a PCR program with a relatively short extension time (30 s), we generated a 420 bp band in wild-type embryos, a 420 bp band and 368 bp band in heterozygous F2 embryos, and a 368 bp band in homozygous F2 mutant embryos (Table 2, Fig. 1a, b). Similarly, for slc41a1, forward and reverse primers flanking the targeted sequence were predicted to produce a 2771 bp band in wild-type embryos. However, with the three-primer reaction, we generated a 498 bp band in wild-type embryos, a 498 bp band and 198 bp band in heterozygous embryos, and a 198 bp band in homozygous mutant embryos (Table 2, Fig. 2a, b).
For example, in the case of arhgap29b, 5 out of 8 embryos had a PCR product of the expected size for the mutated sequence.
In the case of arhgap29b, 30 out of 80 (37.5%) F0 fish were positive in a fin-clip screen and 3 out of 5 such positive individuals screened transmitted the mutation to the next generation. However, we do not routinely screen F0s because the presence of mutations in somatic tissue does not guarantee their presence in the germline.
Anesthetizing solution: Add 1 ml of 1% tricaine to 100 ml of fish water. Prepare it fresh when required. Do not over-anesthetize the fish as this could lead to death. After anesthetization, fish should begin to swim within a minute after placing them back in fish water.
Check that the individual F1s (males and females) have the desired mutation by PCR and sequencing. Cross F1 adults to yield F2 embryos.
In the case of arhgap29b, a cross of two F1 carriers yielded a clutch where about 25% of embryos had a distinctive and consistent phenotype of severe edema and poor blood circulation (Fig. 3). This phenotype matched our expectations for the arhgap29b mutant as Arhgap29 is implicated in lumen formation in blood vessels [32–34]. Disappointingly, however, the animals with this phenotype were not consistently homozygous for the promoter-deletion genotype. This implies the CRISPR/Cas9 reagents for arhgap29b yielded an unintended mutation that in homozygous form caused the observed phenotype. Given this phenotype matched our expectation for an arhgap29b mutant, it is possible that the off-target mutation is within the arhgap29b gene and is not detected by our PCR assay. We have seen a similar pattern of a predicted phenotype not segregating with the targeted mutation in two other instances; these coincidences remain unexplained. To identify the off-target mutation, one option is to perform whole-genome sequencing of embryos with normal and abnormal phenotypes, although efficient use of this approach depends on the use of inbred strains for the mutagenesis and mapping crosses [35]. An alternative approach developed for use in cell lines, called genome-wide, unbiased identification of double-stranded breaks enabled by sequencing (GUIDE-seq), relies on capture of double-stranded oligodeoxynucleotides into all of the double-stranded breaks generated by a given gRNA plus Cas9. One identifies all such oligo-integration sites by sequencing. Later, one subsequently amplifies and sequences all of the sites so identified in a cell line separately mutagenized with that gRNA [36]. The use of GUIDE-seq in zebrafish has not yet been published.
In the case of slc41a1, 5 day postfertilization (dpf) F2 larvae homozygous for the promoter-deletion had normal morphology but lower-than-normal levels of magnesium ion, consistent with larvae injected MO targeting splicing of slc41a1 [37] (Sturgeon and Cornell, in preparation).
The RNA precipitate will form a pellet on the side and bottom of the tube.
After 15 min of incubation start thawing the inactivation reagent on ice.
Leave 4–5 μl of solution in the tube to avoid DNA and buffer salt contamination.
We found the comparative CT method as described by Schmittgen and Livak [38] to be an appropriate analysis method for our experiments.
We detected a significant reduction, although not a complete loss, of arhgap29b mRNA expression in homozygous mutant embryos as compared with wild-type embryos. We expect that the residual expression of arhgap29b expression is of maternal origin. To test this possibility, we are raising homozygous mutant adults to adulthood and will assess arhgap29b expression in maternal-zygotic mutants in the next generation. Embryos that are homozygous for a deletion in the slc41a1 gene did not have detectable slc41a1 transcript. In cDNA prepared from 2 out of the 3 RNA samples from pooled embryos, we did not see any amplification of slc41a1 by real-time PCR even after 40 cycles.
References
- 1.Marsh S et al. (2020) Application of CRISPR/Cas9-mediated genome editing for the treatment of myotonic dystrophy type I. Mol Ther 28(12):2527–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong L et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121):819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaj T (2021) Next-generation CRISPR technologies and their applications in gene and cell therapy. Trends Biotechnol 39(7):692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talbot JC, Amacher SL (2014) A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish 11(6):583–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackburn PR et al. (2013) The CRISPR system—keeping zebrafish gene targeting fresh. Zebrafish 10(1):116–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sertori R et al. (2016) Genome editing in zebrafish: a practical overview. Brief Funct Genomics 15(4):322–330 [DOI] [PubMed] [Google Scholar]
- 7.Hwang WY et al. (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31(3):227–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M et al. (2016) Zebrafish genome engineering using the CRISPR–Cas9 system. Trends Genet 32(12):815–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ota S et al. (2014) Multiple genome modifications by the CRISPR/Cas9 system in zebrafish. Genes Cells 19(7):555–564 [DOI] [PubMed] [Google Scholar]
- 10.Hoshijima K et al. (2019) Highly efficient CRISPR-Cas9-based methods for generating deletion mutations and F0 embryos that lack gene function in zebrafish. Dev Cell 51 (5):645–657.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hisano Y et al. (2015) Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci Rep 5 (1):1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boel A et al. (2018) CRISPR/Cas9-mediated homology-directed repair by ssODNs in zebrafish induces complex mutational patterns resulting from genomic integration of repair-template fragments. Dis Model Mech 11(10): dmm035352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wierson WA et al. (2018) GeneWeld: a method for efficient targeted integration directed by short homology. BioRxiv:431627 [Google Scholar]
- 14.Gagnon JA et al. (2014) Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9(5):e98186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burger A et al. (2016) Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 143 (11):2025–2037 [DOI] [PubMed] [Google Scholar]
- 16.Johnson MJ et al. (2018) Engineering of primary human B cells with CRISPR/Cas9 targeted nuclease. Sci Rep 8(1):1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shapiro J et al. (2020) Increasing CRISPR efficiency and measuring its specificity in HSPCs using a clinically relevant system. Mol Ther Methods Clin Dev 17:1097–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kok FO et al. (2015) Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell 32(1):97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawson ND (2016) Reverse genetics in zebrafish: mutants, morphants, and moving forward. Trends Cell Biol 26(2):77–79 [DOI] [PubMed] [Google Scholar]
- 20.El-Brolosy MA et al. (2019) Genetic compensation triggered by mutant mRNA degradation. Nature 568(7751):193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leslie EJ et al. (2012) Expression and mutation analyses implicate ARHGAP29 as the etiologic gene for the cleft lip with or without cleft palate locus identified by genome-wide association on chromosome 1p22. Birth Defects Res A Clin Mol Teratol 94(11):934–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letra A et al. (2014) Further evidence suggesting a role for variation in ARHGAP29 variants in nonsyndromic cleft lip/palate. Birth Defects Res A Clin Mol Teratol 100(9):679–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H et al. (2016) Exome sequencing provides additional evidence for the involvement of ARHGAP29 in Mendelian orofacial clefting and extends the phenotypic spectrum to isolated cleft palate. Birth Defects Res A Clin Mol Teratol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sturgeon M, Perry Wu RC (2016) SLC41A1 and TRPM7 in magnesium homeostasis and genetic risk for Parkinson’s disease. J Neurol Neuromed 1(9):23. [PMC free article] [PubMed] [Google Scholar]
- 25.Heintzman ND et al. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39(3):311–318 [DOI] [PubMed] [Google Scholar]
- 26.Bogdanović O et al. (2013) The developmental epigenomics toolbox: ChIP-seq and MethylCap-seq profiling of early zebrafish embryos. Methods 62(3):207–215 [DOI] [PubMed] [Google Scholar]
- 27.Liu H et al. (2020) Analysis of zebrafish periderm enhancers facilitates identification of a regulatory variant near human KRT8/18. elife 9:e51325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carninci P et al. (2006) Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38(6):626–635 [DOI] [PubMed] [Google Scholar]
- 29.Kimura K et al. (2006) Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res 16(1):55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jinek M et al. (2012) A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity.Science 337 (6096):816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaFave MC et al. (2014) A defined zebrafish line for high-throughput genetics and genomics: NHGRI-1. Genetics 198(1):167–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Post A et al. (2013) Rasip1 mediates Rap1 regulation of Rho in endothelial barrier function through ArhGAP29. Proc Natl Acad Sci 110 (28):11427–11432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu K, Cleaver O (2011) Tubulogenesis during blood vessel formation. In: Seminars in cell & developmental biology. Elsevier; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu K et al. (2011) Blood vessel tubulogenesis requires Rasip1 regulation of GTPase signaling. Dev Cell 20(4):526–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolman MA et al. (2015) A genome-wide screen identifies PAPP-AA-mediated IGFR signaling as a novel regulator of habituation learning. Neuron 85(6):1200–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai SQ et al. (2015) GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33 (2):187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arjona FJ et al. (2019) SLC41A1 is essential for magnesium homeostasis in vivo. Pflügers Arch Eur J Physiol 471(6):845–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C T method. Nat Protoc 3(6):1101. [DOI] [PubMed] [Google Scholar]