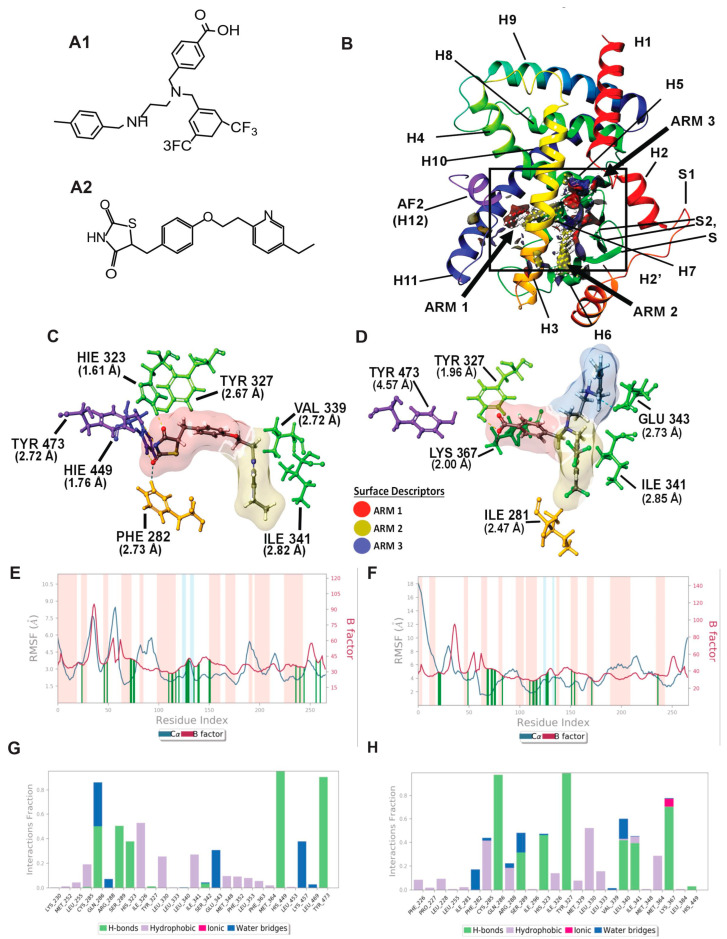
Figure 2.
Molecular modeling: In silico modeling of PPARγ interactions between Pio (PPARγ agonist) and AU9. (A) Chemical structure of AU9 (A1) and Pio (A2). (B) Site Map analysis of PPARγ LBD PDB: 5Y2O. Hydrophobic surface map (yellow), hydrogen-bond donor surface map (blue), and hydrogen-bond acceptor surface map (red). α-helices and β-sheets labeled H1–H12 and S1–S4, respectively, from N-terminus to C-terminus. Y-shaped ligand binding pocket where arm 1 contains the AF2 domain, arm 2 is the entrance site, and arm 3 is a secondary ligand binding pocket. (C) Lowest energy conformation for Pio and amino acid binding interactions with distances (angstroms). Pio forms a hydrogen bond network to the AF2 domain indicative of a full agonist. (D) AU9 lowest energy conformation and amino acid binding interactions with distances. AU9 avoids a key interaction at TYR473 as its branched molecular structure inhibits extension further into the AF2 as compared to Pio. (E,F) Molecular dynamics root-mean-square fluctuation (RMSF) plots. Where the blue graph represents the ligand induced α-carbon fluctuation overlayed with red graph experimental b-factor, α-helices and β-sheets are shaded red and blue, respectively, from N-terminus to C-terminus. Vertical green bars indicate ligand–residue contacts. (E) Pio displays ligand-induced stabilization of the AF2 domain (residue index > 250) represented by a decrease in the RMSF as compared to the b-factor plot. (F) AU9 displays ligand induced stabilization in the global protein structure characterized by a reduction in RMSF as compared to the b-factor; however, avoidance of AF2 interactions cause a greater fluctuation in both the N-terminus and C-terminus indicating partial activity relative to a full agonist. (G,H) Protein interaction diagram categorized by the fraction and type of interactions maintained through the course of simulation. (G) Pio maintains hydrogen bonds to the AF2 residue TYR 473 with supporting interactions at residues HIS 449, HIS 323, SER 289, and GLN 286 as the major contribution of ligand-protein contacts. (H) AU9 maintains only minimal contact to the AF2 supporting residue HIS 449, thus contributing to the increased RMSF values observed in this region of the protein.