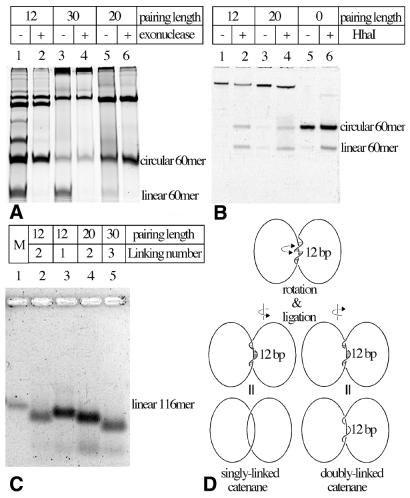
Figure 2.
Gel-electrophoretic analysis of ligation reactions leading to the formation of various catenanes. (A) Exonuclease eliminates linear single-stranded DNA molecules and reveals DNA species that are likely to be catenanes. Notice that 12 nt-long complementary sequences between the two 60mers (annealing of these sequences is required for the circularization of the ‘second’ 60mer, but can also lead to formation of linear multimers composed of these 60mers) result in frequent formation of dimeric and trimeric linear species, which are therefore exonuclease sensitive. Longer complementary sequences eliminate the formation of these dimeric and trimeric linear molecules but promote the formation of species with high molecular weight (see the top of the gel). (B) Partial restriction cleavage of one of the 60mers unambiguously demonstrates the catenated nature of DNA species that we expected to be catenated. In lanes 2 and 4 notice the equal intensity of circular and linear 60mers released from gel-purified slowly migrating species. In lane 6 circular 60mers are partially cut by HhaI and get partially converted to linear 60mers. (C) In alkaline agarose gels catenanes separate according to their expected linking number. M, marker lane containing linear 116mer. (D) Formation of singly and doubly linked catenanes in the case of 12 nt-long complementarity between the two rings. With these substrates the catenanes with linking number 1 (left) and 2 (right) show small energy difference and therefore form with similar probability. (A and B) PAA-denaturing gels with urea and (C) alkaline agarose NuSieve gel 4%.