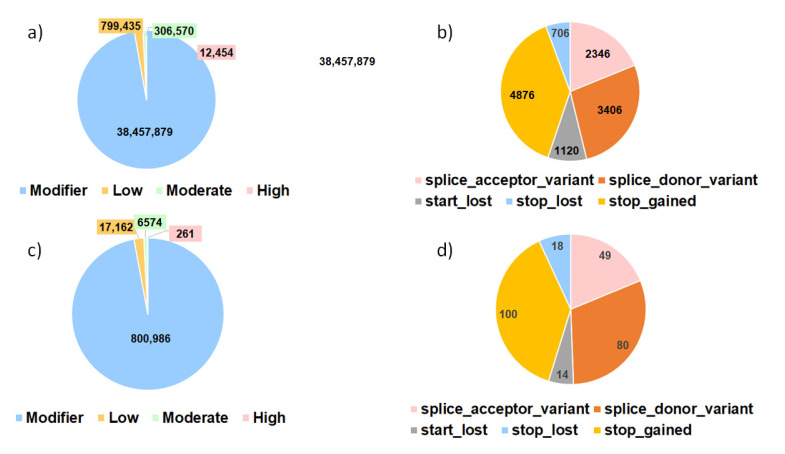
Figure 5.
Proportion of variants detected (%) in the variant effect analyses for all the SNPs detected in the variant calling (a,b) and the variants identified in the detected outlier regions from the FST analyses (c,d). (a,c) describe all the variants divided according to their predicted impact. Modifier: usually non-coding variants or variants affecting non-coding genes, where predictions are difficult or there is no evidence of impact. Moderate: a non-destructive variant that can change protein effectiveness. Low: assumed to be mostly harmless or unlikely to change protein behavior. High: the variant is assumed to have high (disruptive) impact in the protein, probably causing protein truncation, loss of function, or triggering nonsense mediated decay. (b,d) include only the proportions of the different predicted variations of the SNPs with high impact.