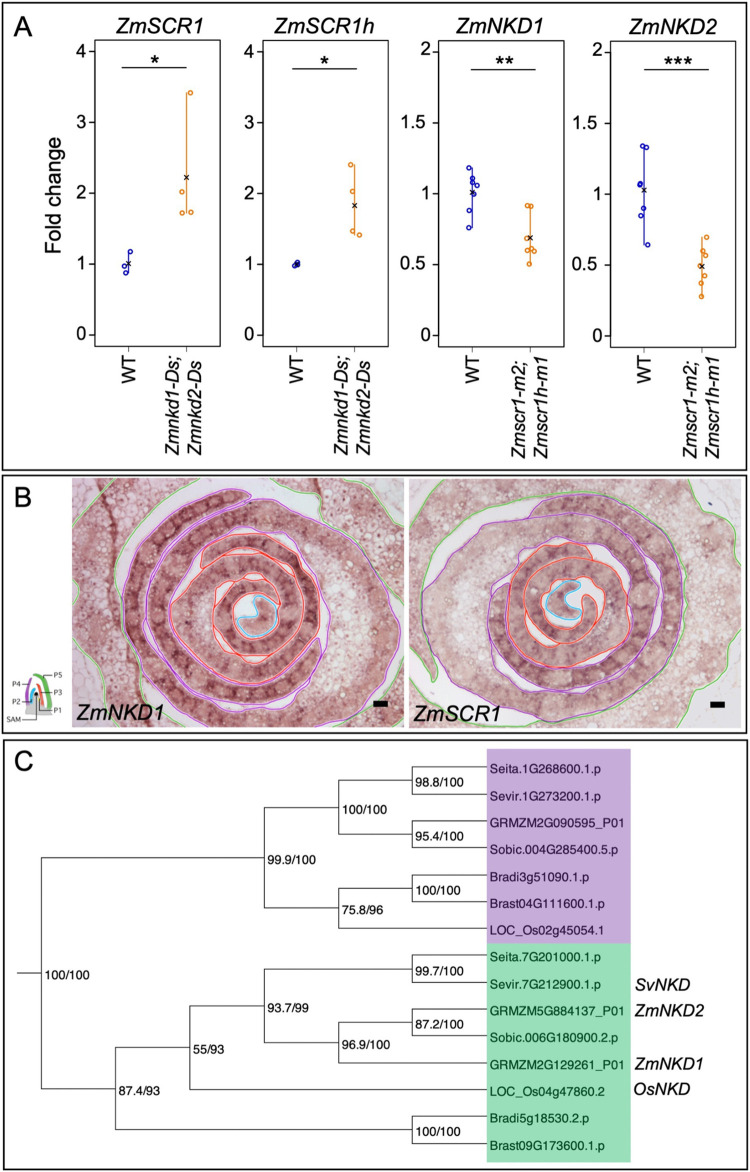
Fig 3. NKD and SCR transcripts accumulate in the same spatial domain with levels determined by a feedback loop.
A) Quantitative RT-PCR of ZmSCR1 and ZmSCR2 transcripts in the Zmnkd1-Ds;Zmnkd2-Ds mutant, and ZmNKD1 and ZmNKD2 in the Zmscr1-m2;Zmscr1h-m2 mutant. Open circles are individual biological replicates, black crosses indicate the mean for each genotype. Statistical significance as calculated on log-transformed fold change data by Welch’s t-test (two-tailed) indicated above each plot: *P≤0.05; **P≤0.01; ***P≤0.001 (raw data in S1 Table). B) In-situ hybridization to ZmNKD1 and ZmSCR1 in maize wild-type B73 apices. In each image the P2 primordium is outlined in blue, P3 in red, P4 in purple and P5 in green, as indicated in the adjacent cartoon diagram. Darker purple signal represents successful hybridization to each transcript of interest. Scale bars: 50μm. C) Maximum likelihood phylogeny of the NKD genes in monocots. The NKD clade is highlighted in green, and the adjacent monocot clade in purple. Bootstrap values are displayed at each branch of the phylogeny.