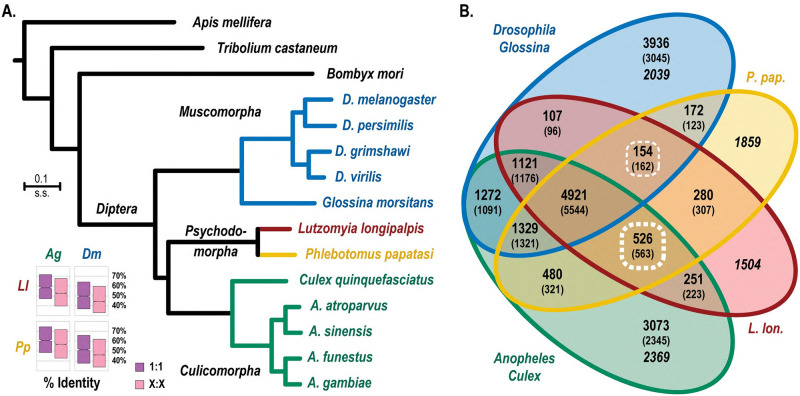
Fig 2. Molecular species phylogeny and ortholog sharing.
(A) The quantitative maximum likelihood species phylogeny computed from the concatenated superalignment of 1,627 orthologous protein-coding genes places the sand flies (Psychodomorpha) as a sister group to the mosquitoes (Culicomorpha) rather than the flies (Muscomorpha), with all branches showing 100% bootstrap support. The Culicomorpha are represented by four Anopheles mosquito species and Culex quinquefasciatus and the Muscomorpha include four Drosophila fruit fly species and the tsetse fly, G. morsitans. Outgroup species represent Lepidoptera (Bombyx mori), Coleoptera (T. castaneum), Hymenoptera (Apis mellifera), and the phylogeny is rooted with the phthirapteran human body louse, Pe. humanus. The inset boxplots show that single-copy (1:1) and multi-copy (X:X) ortholog amino acid percent identity is higher between each sand fly (Ll, Lu. longipalpis; Pp, Ph. papatasi) and An. gambiae (Ag) than D. melanogaster (Dm). Boxplots show median values with boxes extending to the first and third quartiles of the distributions. (B) The Venn diagram summarizes the numbers of orthologous groups and mean number of genes per species (in parentheses) shared among the two sand flies (L. lon., Lu. longipalpis; P. pap., Ph. papatasi) and/or the Culicomorpha and/or the Muscomorpha. Analysis of ortholog sharing shows that the sand flies share more than three times as many orthologous groups exclusively with the Culicomorpha (Anopheles and Culex) compared to the Muscomorpha (Drosophila, Glossina) (subsets highlighted with thin and thick dashed lines). Numbers of unique genes are in italics. Colors in panel A and panel B match species and sets of species analyzed.