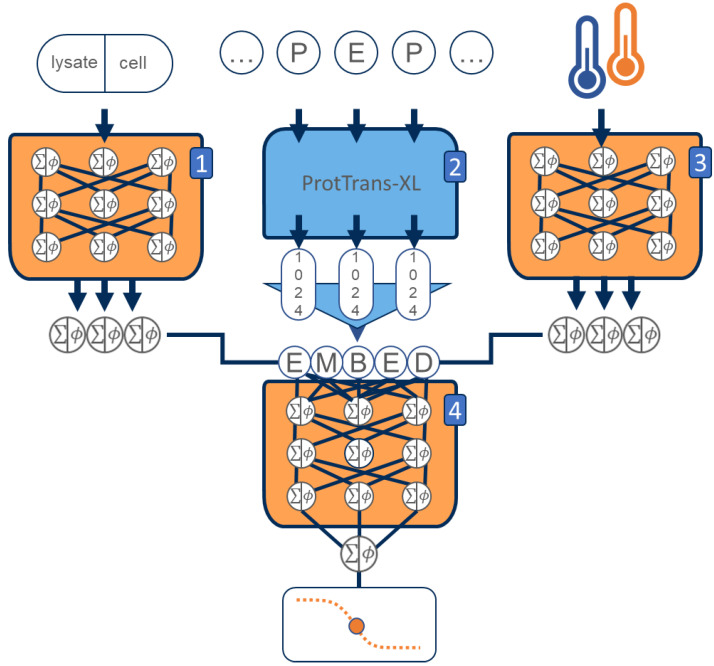
Figure 2.
Schematic overview of the deep learning architecture DeepSTABp trained to predict protein melting temperatures. The proposed model is based on four different artificial network blocks. The first three blocks are responsible of creating an embedding of the protein query based on the input features, (1) type of experimental condition used in the thermal proteome profiling experiment, (2) the protein amino acid sequence and (3) the organism growth temperature. While Blocks 1 and 3 use small multilayer perceptrons (MLP), Block 3 consists of the pretrained transformer-based model ProtTrans-XL, followed by a mean pooling layer. The output vectors of the first blocks are concatenated and then used as inputs for a (4) final MLP block, whose output is the predicted protein Tm.