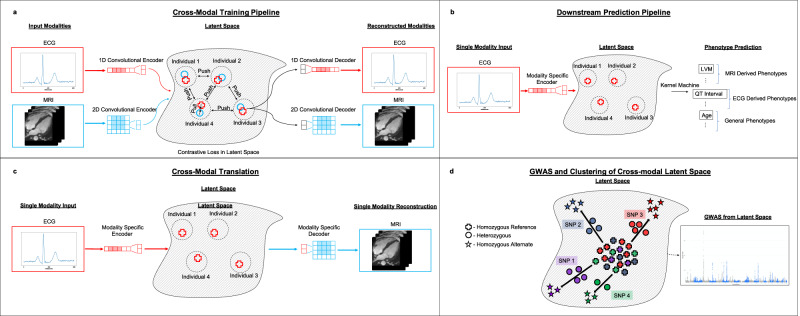
Fig. 1. An overview of our cross-modal autoencoder framework for integrating cardiovascular data modalities.
Our model is trained on ECG and cardiac MRI pairs from the UK Biobank. a A visualization of our training pipeline. Modality-specific encoders map data modalities into a shared latent space in which a contrastive loss is used to enforce the constraint that paired samples are embedded nearby and further apart from other samples. Modality specific decoders are then used to reconstruct modalities from points in the latent space. b Learned cross-modal representations are used for downstream phenotype prediction tasks by training a supervised learning model (e.g., a kernel machine) on the latent representations. c Our framework enables translation between modalities: ECGs can be translated to corresponding MRIs and vice-versa. d The learned cross-modal representations can be used to understand genotype-phenotype maps in the absence of labelled phenotypes by performing a GWAS in the cross-model latent space and clustering SNPs via their signatures (i.e., the vector in latent space oriented from homozygous reference to the mean of heterozygous and homozygous alternate); SNPs 1 and 4 have similar signatures in the latent space and thus similar phenotypic effects.