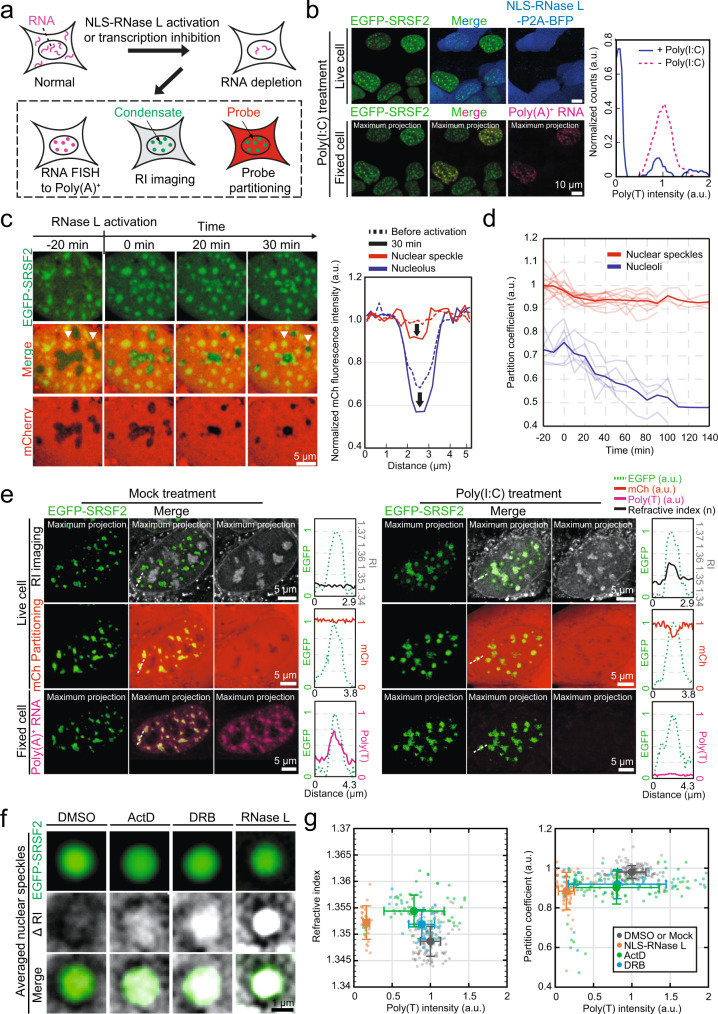
Fig. 4. RNA tunes the biomolecular density of intracellular condensates.
a Schematics of RNA depletion experiments. After activating NLS-RNase L by poly(I:C) treatment or treating cells with either DRB or ActD, a series of quantitative characterizations were conducted. b (Left) Representative images of U2OS cells stably expressing EGFP-SRSF2 and NLS-RNase L-P2A-BFP after poly(I:C) treatment. After live-cell images were collected, poly-dT RNA FISH images were acquired for the same cells. (Right) Distribution of poly-dT intensities within individual nuclear speckles. Only cells expressing both constructs were included. The normalized counts were averaged with a sliding window of 0.2 (a.u.). n = 146 (mock) and 119 (poly(I:C)) c (Left) Time-lapse images of a live U2OS cell stably expressing EGFP-SRSF2, mCherry, and NLS-RNase L after poly(I:C) treatment. Time 0 is defined when the nuclear speckle morphology begins to change. (Right) Normalized intensity profiles for mCherry across either a nuclear speckle or a nucleolus labeled with arrowheads in (c; Left). d Temporal changes of the partition coefficients of mCherry for nuclear speckles or nucleoli in U2OS cells after NLS-RNase L activation. Time is defined as in (c). The bold lines denote average values. n = 14 (nuclear speckles) and 6 (nucleoli). e Representative images of U2OS cells expressing EGFP-SRSF2, mCherry, and NLS-RNase L-P2A-BFP under mock treatment (Left) or poly(I:C) treatment (Right). After imaging live-cells, poly-dT RNA FISH images were acquired for the same cells. Normalized intensity profiles of EGFP-SRSF2 (green), RI (gray), mCh (red), and poly-dT (pink) along white dashed lines are shown. RI images are adjusted to the range of 1.34–1.37. f Averaged RI and fluorescence images of nuclear speckles for each RNA depletion condition. Individual images of nuclear speckles (same datasets as in g; Left) were center-aligned using fluorescence signals before averaging. ΔRI images were obtained by subtracting the minimum RI pixel value of each averaged RI image and adjusted to the range of 0–0.0025. g For individual nuclear speckles, either refractive indices (Left) or partition coefficients of mCherry (Right) were plotted against poly-dT intensities. Each experimental condition is color-coded: gray (DMSO or Mock), orange (NLS-RNase L), green (ActD), and blue (DRB). Data: center dot = mean; whiskers = [mean-std, mean+std].