Summary
The inner ear of humans and large animals is embedded in a thick and dense bone that makes dissection challenging. Here, we present a protocol that enables three-dimensional (3D) characterization of intact inner ears from large-animal models. We describe steps for decalcifying bone, using solvents to remove color and lipids, and imaging tissues in 3D using confocal and light sheet microscopy. We then detail a pipeline to count hair cells in antibody-stained and 3D imaged cochleae using open-source software.
For complete details on the use and execution of this protocol, please refer to (Moatti et al., 2022).1
Subject areas: Microscopy, Biotechnology and Bioengineering
Graphical abstract

Highlights
-
•
This protocol supports the investigation of the 3D cytoarchitecture of the inner ear
-
•
Removal of Ca, lipids, and pigments to improve light penetration by several mm
-
•
Tissue clearing increases antibody diffusion to analyze larger animal inner ears
-
•
Hair cell counting at 3D subcellular resolution using open-source software
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
The inner ear of humans and large animals is embedded in a thick and dense bone that makes dissection challenging. Here, we present a protocol that enables three-dimensional (3D) characterization of intact inner ears from large-animal models. We describe steps for decalcifying bone, using solvents to remove color and lipids, and imaging tissues in 3D using confocal and light sheet microscopy. We then detail a pipeline to count hair cells in antibody-stained and 3D imaged cochleae using open-source software.
Before you begin
The protocol below describes the specific steps for tissue clearing, labeling, imaging, and quantifying the inner ear of fetus and young – juvenile pigs. We show that this protocol also works well with non-human primate inner ear (NHP; African green monkey). Therefore, we believe that this method can be used for other species with dense bone such as rabbits, gerbils, guinea pigs, and humans, although modifications might be required.
Institutional permissions
All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at North Carolina State University, following the standards of the National Institute of Health and Committee on Care and Use of Laboratory Animals. If you are ordering animals for tissue collection, you might need to acquire institutional permission.
Key resources table
| REAGENTS or RESOURCES | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-MYO7a (1:200 dilution) | Proteus | 25-6790 |
| Rabbit anti-PGP9.5 Polyclonal (1:200 dilution) | Proteintech | 14730-1-AP |
| Cy™3 AffiniPure Donkey Anti-Rabbit IgG (H+L) (1:250 dilution) | Millipore | 711-165-152 |
| Chemicals, peptides, and recombinant proteins | ||
| Phosphate buffered saline Powder--PBS | Sigma-Aldrich | P3813-5X10PAK |
| Triton-X100 | Sigma-Aldrich | X100-500ML |
| Tween-20 | Sigma-Aldrich | P9416-100ML |
| Dimethyl Sulfoxide--DMSO | Fisher Scientific | D128-1 |
| Donkey Serum | Sigma-Aldrich | D9663-10ML |
| Heparin | Sigma-Aldrich | H3393-100KU |
| Methanol--MeOH | Fisher Scientific | A412SK-4 |
| Hydrogen Peroxide 30% | Sigma-Aldrich | 216763-500ML |
| Dibenzyl Ether--DBE | Sigma-Aldrich | 108014-3KG |
| 32% Paraformaldehyde--PFA aqueous solution | Electron Microscopy Sciences | RT 15714-S |
| Ethylenediaminetetraacetic acid-EDTA | Sigma-Aldrich | EDS-500G |
| Sucrose | Sigma-Aldrich | S0389-1KG |
| Sodium Hydroxide--NaOH | Sigma-Aldrich | S8045 |
| Deoxycholate | Sigma-Aldrich | D6750-25G |
| Ethylenediaminetetraacetic acid-disodium-EDTA-Na2 | Sigma-Aldrich | E5134-500G |
| Dichloromethane--DCM | Sigma-Aldrich | 270997-1L |
| Sodium Azide (optional) | Sigma-Aldrich | 26628-22-8 |
| Silicon Glue (optional) | Amazon | Gorilla |
| Nail enamel (optional) | Amazon | N/A |
| Deposited data | ||
| Quantified raw data of LGR5 expression | Mendeley Data | https://doi.org/10.17632/vdw3757cxv.3 |
| Experimental models: Organisms/strains | ||
| Pigs, both sexes, postnatal day 0 and 60 | NCSU Swine Education Unit farm | Wild type (Yorkshire) |
| African Green Monkey, male, adult 5+ years | NCSU CVM, Dr. Cheng lab | Wild type |
| Software and algorithms | ||
| IMARIS | Oxford Instruments | https://imaris.oxinst.com/packages |
| Ilastik | Berg, S et al. | https://pubmed.ncbi.nlm.nih.gov/31570887/ |
| Terastitcher | Bria, A et al. | https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-13-316 |
| ImageJ | GitHub | https://github.com/imagej |
| Prism 9.2 graphpad | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Biorender | BioRender | https://biorender.com/ |
| Other | ||
| Bone cutting forceps | Fisher Scientific | Liston Integra Miltex |
| Bone Nipper | Fine Science Tools | Angled, 11 cm |
| Bone Stryker | N/A | N/A |
| Standard Dissecting scissor | Fisher Scientific | N/A |
| Standard tweezer | Fisher Scientific | N/A |
| Scalpel #10 | N/A | N/A |
| Shaker 4C (optional) | VWR | 12620-938 |
| Shaker RT | VWR | 10127-872 |
| Incubating Shaker | VWR | 444-0762 |
| Eppendorf tubes | 5 and 50 mL | |
| Parafilm | N/A | N/A |
| Aluminum foil | N/A | N/A |
| Needle (Optional) | 30 ga | |
| Perfusion pump (Optional) | Instech | P720/37K |
| Silicon tubing (Optional) | Instech | 0.015" ID x 0.78" OD |
| Rat Intrathecal catheter (Optional) | Instech | 32 ga (0.8Fr) PU 18 cm, stylet, 27 ga luer stub.Fits 27 ga |
| Light-sheet microscope | Olympus | XLPLN10XSVMP-2; 10x/NA 0.6 objective |
| Silicon membrane | McMaster-Carr | 87315K71 |
| Confocal microscope | Olympus | FLUOVIEW FV3000; 4x/NA 0.16, 10x/NA 0.75, and 30x Si oil/NA 1.05 Plan Apo objectives |
| iSpacers for confocal imaging (optional) | SUNJin Lab | 0.15, 1, a620nd 3mm |
Alternatives: A confocal microscope can be used instead of a light-sheet microscope. However, the acquisition time might be lengthy, and the sample might photo-bleach. If the confocal microscope is used, we recommend focusing on a specific region of interest that a series of images can be acquired in ∼12–16 h.
Materials and equipment
Fixative solution I
| Reagent | Final concentration | Amount |
|---|---|---|
| PFA (32%) | 1% | 1.56 mL |
| Sucrose | 10% | 5 gr |
| 10 × PBS | 1× | 5 mL |
| ddH2O | N/A | ∼43 mL |
| Total | N/A | 50 mL |
Note: The stock can be stored for up to 1 week at 4°C.
Fixative solution II
| Reagent | Final concentration | Amount |
|---|---|---|
| PFA (32%) | 0.5% | 0.78 mL |
| 10 × PBS | 1× | 5 mL |
| ddH2O | N/A | ∼44 mL |
| Total | N/A | 50 mL |
Note: The stock can be stored for up to 1 week at 4°C.
Decalcification buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| EDTA-Na2 | 350 mM | 61.29 gr |
| NaOH (10 M) | PH of 6.5 | ∼15 mL |
| 10 × PBS | 1× | 50 mL |
| ddH2O | N/A | ∼430 mL |
| Total | N/A | 500 mL |
Note: The stock can be stored at 4°C for 6 months.
Methanol gradients solutions
-
•
20% Methanol solution: add 40 mL Methanol (100%) in 160 mL ddH2O.
-
•
40% Methanol solution: add 80 mL Methanol (100%) in 120 mL ddH2O.
-
•
60% Methanol solution: add 120 mL Methanol (100%) in 80 mL ddH2O.
-
•
80% Methanol solution: add 160 mL Methanol (100%) in 40 mL ddH2O.
-
•
100% Methanol solution.
Note: The stock can be stored for 3 months at 25°C in a safety cabinet. You can store methanol for a longer time if the vial containers are fully sealed and there is no evaporation.
Decolorization buffer (3%): add 5 mL H2O2 (30%) in 50 mL Methanol (100%).
Note: The decolorization buffer needs to be made immediately before use.
Permeabilization buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Triton X-100 | 0.2% | 0.1 mL |
| Deoxycholate | 0.1% | 0.05 gr |
| DMSO | 10% | 5 mL |
| EDTA | 25 mM | 0.36 gr |
| 10 × PBS | 1× | 5 mL |
| NaOH (1 M) | PH of 6.5 | ∼3 mL |
| ddH2O | N/A | ∼37 mL |
| Total | N/A | 50 mL |
Note: The stock can be stored at 4°C for 6 months.
Blocking buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Triton X-100 | 0.2% | 0.1 mL |
| Normal Donkey Serum | 5% | 2.5 mL |
| DMSO | 10% | 5 mL |
| EDTA | 25 mM | 0.36 gr |
| 10 × PBS | 1× | 5 mL |
| NaOH (1 M) | PH of 6.5 | ∼3 mL |
| ddH2O | N/A | ∼34 mL |
| Total | N/A | 50 mL |
Note: The stock can be made without donkey serum and stored at 4°C for 6 months and fresh donkey serum can be added immediately before use.
Antibody buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tween-20 | 0.2% | 0.02 mL |
| Normal Donkey Serum | 5% | 0.5 mL |
| Heparin | 10 μg/mL | 0.1 mg |
| EDTA | 25 mM | 0.07 gr |
| 10 × PBS | 1× | 1 mL |
| NaOH (1 M) | PH of 6.5 | ∼0.5 mL |
| ddH2O | N/A | ∼8 mL |
| Total | N/A | 10 mL |
Note: The stock can be made without donkey serum and stored at 4°C for 6 months and fresh donkey serum can be added immediately before use.
Washing buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tween-20 | 0.2% | 1 mL |
| Heparin | 10 μg/mL | 5 mg |
| EDTA | 25 mM | 3.6 gr |
| 10 × PBS | 1× | 50 mL |
| NaOH (1 M) | PH of 6.5 | ∼25 mL |
| ddH2O | N/A | ∼422 mL |
| Total | N/A | 500 mL |
Note: The stock can be stored at 4°C for 6 months.
Delipidating solution I (66%): add 10 mL DCM (100%) in 5 mL Methanol (100%).
Note: The delipidating solution needs to be made immediately before use.
Delipidating solution II: 50 mL of DCM (100%).
Refractive index--RI matching and Imaging solution: 300 mL DBE (100%).
CRITICAL: Please take precautions in handling sodium azide (toxic), PFA (toxic and carcinogen), EDTA and EDTA disodium powders (toxic), methanol (toxic), DCM (toxic), and DBE (irritable). Avoid inhalation and skin contact and work under a chemical hood.
Step-by-step method details
Dissection
Timing: 10–20 min (for steps 1 and 2)
In this step, dissect the porcine inner ear out of the skull and trim it for tissue clearing. For the large animal model (e.g., pig) we did not perfuse the animal. For small animals, you can perform transcardiac perfusion with 25 mL 1× PBS/50 μg/mL heparin, followed by 25 mL 1× PBS/1% PFA/10% sucrose/50 μg/mL heparin.
-
1.Dissect the inner ear (See Figure 1).
-
a.Separate the head from the body.
-
b.Remove the skin from the mid skull all the way to the back of the neck using a razor blade or scalpel.
-
c.Cut a rectangular window through the bone on top of the skull using a bone stryker.
-
d.Remove the brain and brain stem.
-
e.Locate the inner ears on the lateral sides of the skull.
-
f.Cut right above the inner ear, where the superior margin of the petrous part is located.Note: The petrous part is the border and the most medial part of the temporal bone, with a pyramid shape, and is wedged in the base of the skull.
-
g.Cut the base of the skull in half using the bone stryker. Detach the rest of the inner ear periphery from the skull.
-
h.Push the base of the skull back and separate the lateral parts including the inner ears from the rest of the skull.
-
a.
-
2.Trim the inner ear (see Methods video S1-parts 1, 2, and 3, and Figure 1).
-
a.Trim the excess bone around the inner ear using a bone stryker, the goal is to remove temporal bone while keeping the cochlea intact. Locate the tympanic membrane or middle ear and carefully trim the specimen without damaging the inner ear using bone-cutting forceps.
-
b.Trim the excess bones and make cuts in the spongious mastoid bone using bone-cutting forceps until the apex of the cochlea is found. Remove the loose bone pieces, tissues, and ligaments using a scissor. When you get closer to the inner ear, use the bone nipper to trim. Any temporal bone that was not trimmed could be cleaned later after decalcification.
-
c.Remove the stapes and the round window membrane using a sharp (style 5) tweezer.
-
d.Transfer the inner ear into the fixative solution I in a 50 mL tube for 1–2 h at 25°C.
-
a.
Figure 1.
Dissecting and trimming the porcine inner ear
(A) Expose the inner ear by cutting a rectangular window on top of the skull using a bone stryker, remove the brain and brain stem, and locate the inner ears on the lateral sides of the skull.
(B and C) (B) Locate the superior margin of the petrous part, white arrow in the figure, and cut in parallel to detach the inner ear from the skull as shown in (C).
(D) The tympanic membrane is located in the middle ear side, flip the sample to reveal it.
(E) Trim the spongious mastoid bone using bone-cutting forceps and expose the apex of the cochlea.
(F) Remove the stapes.
(G) Remove the round window membrane.
(H) The dissected and trimmed inner ear top view (up) and side view (down). Related to steps 1 and 2.
Tissue clearing
Timing: 27–33 days (for step 3)
In this step, label the tissue using immunofluorescence, and render the entire inner ear transparent.
-
3.We optimized BoneClear2 tissue-clearing protocol to render the porcine inner ear transparent. However, we believe that this method can also be used with other animal models such as adult mice, guinea pigs, gerbils, rabbits, and non-human primates. To demonstrate the applicability of the method across species, other than pigs, we have cleared a cochlea from an adult African green monkey (See Figure 2).
-
a.Further fix the inner ear in the fixative solution II 12–20 h at 4°C.
-
i.Wash the sample 3 times in 1× PBS in a 50 mL tube for 1 h each.
-
i.
-
b.Decalcify the bone via the decalcification buffer inside a 50 mL tube for 2–10 days based on the age of the animal at 37°C on a shaker with a fresh buffer exchange every 24 h.
-
i.If there is any remaining bone, separate the inner ear from the temporal bone using a bone cutter.
-
i.
-
c.Dehydrate the sample using methanol gradients at 25 oC on a shaker. Please change the solution under the chemical hood using the same 50 mL tube:
-
i.Incubate in 20% methanol for 2 h.
-
ii.Incubate in 40% methanol for 2 h.
-
iii.Incubate in 60% methanol for 2 h.
-
iv.Incubate in 80% methanol for 2 h.
-
v.Incubate in 100% methanol for 2 h, repeat this step, (v), twice.
-
i.
-
d.Decolorize the sample using the decolorization buffer at 4°C 12–20 h on a shaker.
-
e.Rehydrate the sample using methanol gradients at RT on a shaker:
-
i.Incubate in 100% methanol for 2 h.
-
ii.Incubate in 80% methanol for 2 h.
-
iii.Incubate in 60% methanol for 2 h.
-
iv.Incubate in 40% methanol for 2 h.
-
v.Incubate in 20% methanol for 2 h.
-
vi.Incubate in PBS for 2 h.
-
i.
-
f.Immunolabel the samples.
-
i.Place the sample in an a 50 ml tube, fill it with the permeabilization solution, and place it on a shaker with gentle shaking for 12–20 h (37°C).
-
ii.Using the same tube, discard the permeabilization solution, and replace it with the blocking buffer. Place the sample on a shaker for 12–20 h (37°C).
-
iii.Place the sample in an appropriate size tube and immunolabel the sample with primary antibodies that are added to the antibody buffer (Vantibody:Vbuffer = 1:200). The sample is placed at 37°C on a shaker for 5–7 days. For smaller samples and younger animals, use 5 days.Note: Pick the tube size based on the sample size and minimize the solution volume to save immunolabeling reagents. For adult pigs and NHPs, a 5 ml tube should be sufficient. The antibody solution should cover the whole sample. For example, for an adult porcine inner ear, a 1.5–2 mL solution is sufficient to cover the sample. Cover the cap with parafilm to avoid leakage or evaporation. We used the same timing for all antibodies tested in this experiment (Please see the troubleshooting section), however, the optimization of the incubation time with the primary antibody might be required.
-
iv.Wash the sample with the washing buffer at 37°C on a shaker for 24 h with a fresh buffer change every 6 h. If exchanging the washing buffer every 6 h is not feasible, repeat four times with a minimum of 6 h each.
-
v.Immunolabel the sample with secondary antibodies in the antibody buffer (Vantibody:Vbuffer = 1:250) at 37°C on the shaker for 4–5 days.Note: We have identified that Cy3 and AF647 work well with this technique. Pick your primary and secondary antibodies accordingly. Please note, there will be an extensive auto-fluorescence signal in the 488 nm channel. Cover the cap with parafilm to avoid the leakage.
-
vi.Cover the tubes with aluminum foil from this step onward.
-
vii.Wash the sample with the washing buffer at 37°C on a shaker for 48 h with fresh buffer change every 6 h. If exchanging the washing buffer every 6 h is not feasible, repeat eight times with a minimum of 6 h each.
-
i.
-
g.Dehydrate the sample using methanol gradients using a 50 mL tube at 25 oC on a shaker:
-
i.Incubate in 20% methanol for 2 h.
-
ii.Incubate in 40% methanol for 2 h.
-
iii.Incubate in 60% methanol for 2 h.
-
iv.Incubate in 80% methanol for 2 h.
-
v.Incubate in 100% methanol for 2 h two times (2×).
-
i.
-
h.Delipidate at 25 oC on a shaker.
-
i.Incubate in the delipidating solution I for 2 h 2×.
-
ii.Incubate in the delipidating solution II for 0.5 h four times (4×).
-
i.
-
i.RI-match the sample at 25 oC on a shaker.
-
i.Incubate in RI matching solution in a 50 mL tube at 25oC for 24 h three times (3×).Note: Make sure the DBE, RI matching solution, is filled all the way and minimize the air gap on top. DBE oxidizes in the air to form peroxides with different properties which affect the clarity of the tissue. Handle the sample very gently as any slight squeezing can introduce bubbles inside the sample.Optional: To speed up the immunolabeling process (up to three times) a perfusion pump can be used. First, a perfusion chamber that circulates the antibody solution in an Eppendorf tube is built. Two holes are drilled on top of a 5 ml Eppendorf tube that will be used during the staining process. The hole diameter should be matching the microfluidics tube diameter so there is a slight resistance to moving the microfluidics tubes through the holes. Then, insert the inlet and outlet microfluidic tubes through the holes until they are well below the fluid level in the 5 ml tube. Add the antibody buffer that includes the antibodies to the 5 ml Eppendorf tube, and run the pump to fill the microfluidic tubes with the antibody buffer. Make sure there is no bubble formed inside the tubes. Place the sample within, making sure that the solution covers the sample. Position the inner ear so the round window can be accessed. Connect an intrathecal catheter (cut it short if needed) to the tubing inlet, adjust its length inside the tube, and run the pump to fill the catheter with the antibody buffer. Gently insert the catheter into the round window. Then slowly close the tube cap. Please note, do not push the catheter too deep into the round window as it may damage the cochlear fine structure in the base. The perfused solution will exit from the oval window (make sure the stapes and the oval window membrane were removed) into the 5 ml reservoir at 37°C inside the incubator. Make sure that the outlet tube is inside the solution as well, and shaking is not required. Cover the cap including holes and tubes with parafilm and flow the solution using the perfusion pump (flow rate of 1.9 μL/min). Using this chamber, we were able to decrease the length of the immunolabeling time from 7 to 2 days.5 Please note that if perfused too fast, the flow could damage the delicate organs inside the inner ear.
 Pause point: If needed, recommended short and long pause points will be after the completion of b-i or e-vi steps. The sample can be stored in PBS (RT) for a short term (one-two week) or in PBS/0.1% sodium azide for long-term storage at 4°C. Store the samples in a dark space. We tested additional short pause points (one week), after f-iv and f-vii steps and we did not observe any noticeable effects.
Pause point: If needed, recommended short and long pause points will be after the completion of b-i or e-vi steps. The sample can be stored in PBS (RT) for a short term (one-two week) or in PBS/0.1% sodium azide for long-term storage at 4°C. Store the samples in a dark space. We tested additional short pause points (one week), after f-iv and f-vii steps and we did not observe any noticeable effects.
-
i.
-
a.
Figure 2.
Expected results
Images before (left) and after (right) the tissue-clearing protocol. The pig and non-human primate (NHP; African green monkey) inner ears are shown, related to step 3.
Imaging
Timing: ∼10 h (for step 4)
Timing: ∼3 days (for step 5)
In this step, the whole tissue will be imaged.
-
4.Light-sheet microscopy. Although our setup is home-built,5,6,7 general guidelines for other setups still apply.
-
a.Mount the sample inside the imaging chamber.Note: Use a needle that is positioned away from the region of interest and consider the trajectory of the illumination beam. The needle should not block the illumination beam. The sample should be mounted in an orientation that the illumination beam will travel a minimal distance in the decalcified bone, which still scatters and absorbs light. Mount the sample in the orientation that lays the desired region of interest in the XY plane. Please consider that the objective lens axial resolution is always lower than the spatial resolution, i.e., XY resolution is higher than Z. If using a needle (30 ga) to mount the sample, make sure the needle is sharp. Insert the needle when the sample is mounted in DBE to prevent bubble formation. Bubbles are detrimental to the image quality and create blurriness. Typically, top-view or side-view mounting positions are used. For example, for the cochlea, in the top view, the top of the apex is facing the detection axis (almost parallel) while in the side-view, the lateral side of the cochlea is facing the detection axis (see Figure 3).Alternatives: Other techniques to mount the sample are possible, such as embedding the sample in 0.8% agarose block. The agarose embedding should be done after the step g and before the step h of tissue clearing. If you choose to do so, make sure that the whole tissue is embedded in agarose. The positioning in agarose needs to be adjusted based on the desired orientation. In general, for our purposes, the mounting via a needle was the most convenient mounting technique.
-
b.Fill the chamber with DBE imaging solution; the imaging solution should completely cover the detection objective lens and sample.Note: A silicone membrane should be used to prevent the potential damage to the detection objective when immersed in 100% DBE, imaging solution.5 Create a small hole in the membrane (for example via a biospy punch) and stretch the membrane over the objective tightly where the hole is in front of the lens. Also, make sure to remove the imaging solution from the chamber as soon as the imaging is completed.
-
c.Leave the sample in the immersion chamber for 30 min before imaging.
-
d.Observe if the sample orientation is optimal in the microscope field of view and make adjustment if needed. This step can be tedious. Please see note for (4a) for more details.
-
e.If using an adaptive light sheet microscope, start the calibration process.5Note: A porcine cochlea typically occupies a volume of 6 × 6 × 6 mm3, and in our light-sheet microscope, this corresponds to 5 × 5 tiles.
-
f.Start imaging.Note: We imaged the sample with a pixel size of 0.65 × 0.65 μm2. The sampling along the axial direction (z) can be 5, 10, or even 15 μm depending on the biological application. We used 560, and 640 nm lasers to excite the different fluorescence channels in our samples and 488 nm for autofluorescence, which provides structural information.Optional: For custom-built light-sheet microscopes, use Galvo X and Y mirrors to move the detection objective relative to the light-sheet position while imaging.5 Slight variations in the index of refraction of the tissue versus the imaging media change the focal distance of the detection objective and prevent it from overlapping with the illumination beam, which leads to blurred images. The ability to move the detection objective relative to the light-sheet position while imaging (adaptive) can fix that problem and improve the image quality and contrast.
-
a.
-
5.Confocal microscopy. The inner ear can be imaged using a confocal microscope if a light-sheet microscope is not available.Note: The imaging time will be ten times longer in comparison with light-sheet microscopy when acquiring the images at similar imaging configurations. Therefore, it is recommended to focus on smaller volumes of interest.
-
a.Construct an imaging chamber using iSpacer. Stick the iSpacer on a glass slide and use the silicon glue to seal the glass slide and the iSpacer. Let it dry for 30 min.Note: Select the proper iSpacer thickness based on the sample height or thickness. Selecting an iSpacer that is too thick limits your imaging depth, as the cochlea tends to sink to the bottom of the chamber, and the objective lens working distance is limited. Hence, select an iSpacer that is just slightly thicker than the sample.
-
b.Mount the sample inside the iSpacer and fill the iSpacer with DBE.Note: The area of interest within the tissue should directly face the objective lens. If need be, the sample can be glued to the glass using the super glue to adjust the top/side-view, before adding the imaging solution. Please make sure the glue is dried out (30 s) before adding the imaging solution. The imaging solution should completely fill the iSpacer with a slight convex surface formation.
-
c.Remove the sticker from the top of the iSpacer and place a thin cover glass gently on top.Note: Place the cover glass gradually from one side to minimize bubble formation.
-
d.Clean the residue of DBE, use the nail enamel to seal the cover glass/iSpacer interface, and let it dry for 5–10 min.
-
e.Mount the slide on the confocal stage and start imaging.Note: Change the confocal stage if needed to accommodate larger samples.
-
f.If the desired volume is located at the bottom of the chamber, the sample should be flipped or remounted.Note: Use fresh DBE and a new iSpacer to remount the tissue. The lengthy imaging will cause bleaching and bubble formation in the tissue. It is advised to minimize the imaging volume to the area of interest initially.
-
a.
Figure 3.
Typical mounting orientations being used for the cochlear imaging using the light-sheet microscopy
In this schematic, the immersion chamber is viewed from above. The “side-view” and “top-view” mounting orientations are shown, related to step 4.
Expected outcomes
By the end of this process, the intact inner ear sample is imaged, resulting in raw data files of about 200–300 GB (only the cochlea section) for a volume of 6 × 6 × 6 mm3 (axial or Z spacing of 10 μm). Figure 4 shows side-view images of cochleae obtained from an NHP (Figure 4A) and a pig (Figure 4B and Methods video S2). Please note that the porcine cochlea is imaged slightly off from the perfect side-view. Both tissues are cleared with outstanding quality and imaged with subcellular resolution. Figure 5 and Methods video S3 also show the expected resolution of the imaged inner ear after whole-mount top-view imaging.
Figure 4.
Expected imaging results after tissue clearing of spiral ganglion neurons in NHP and pig cochlea
(A) A section out of a 3D volume of NHP cochlea imaged with a light-sheet microscope. In this image, the soma of individual neurons is clearly visible based on their strong autofluorescence in the 488 nm channel. Please note, during the sample processing, part of the neuron bundle is removed.
(B) A 3D reconstruction of the cochlea from a P0 pig and a single section imaged via a confocal microscope. The porcine cochlea was stained against PGP9.5. Related to steps 4 and 5.
Figure 5.
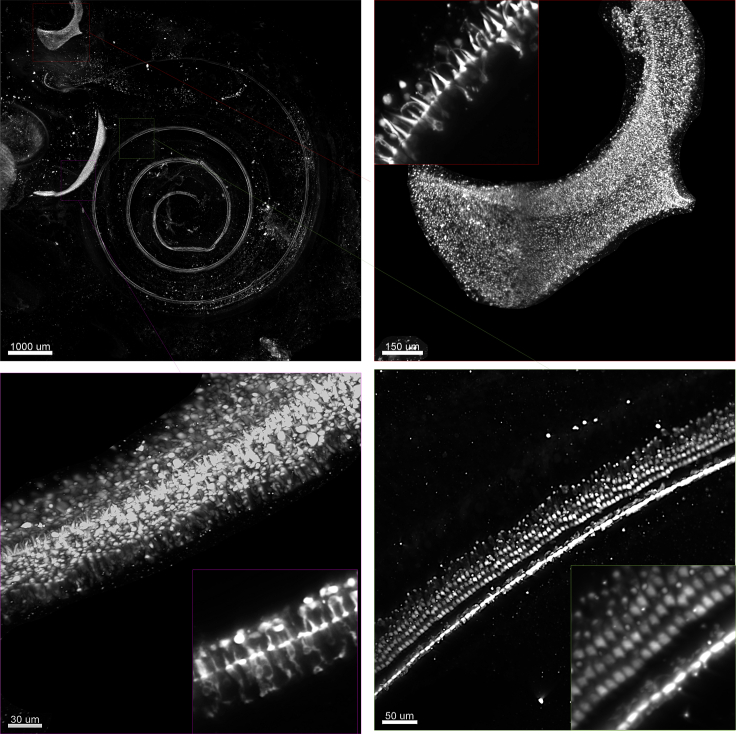
A 3D reconstruction with a subcellular resolution of sensory organs in the inner ear
A P0 porcine inner ear was stained to detect hair cells using anti-MYO7a. The volume was captured using a light-sheet microscope. The zoomed-in areas show subcellular features within the hair cells in the sensory organs of the saccule (purple box), the posterior crista (red box), and the cochlea (green box). Single and zoomed-in slices within the 3D volume are also shown, related to step 4.
Three sensory organs are shown: the saccule, posterior crista, and cochlea, related to step 4.
Quantification and statistical analysis
Timing: ∼2 days (for steps 1 to 5)
In this step, the images will be visualized, processed, and analyzed using image processing tools and machine learning algorithms.
-
1.Data Visualization.
-
a.The stitching and fusion process of multiple imaging tiles can be performed with TeraStitcher.8 Terastitcher has a graphical user interface (GUI) to facilitate the stitching process.Note: Normally the sample is too large to fit into one single imaging tile (1.33 × 1.33 mm2 in our custom-built light-sheet microscope), so multiple overlapping (15%) tiles are captured and stitched to reconstruct the entire dataset.
-
i.Import the volume under the import tab. The first step is to select the path where the two-level hierarchy folder of the volume is stored and specify the “I/O plugin”. Then, specify the physical coordinate system used by the acquisition system, and the pixel size in each dimension. Start the importing process after the above-mentioned acquisition parameters have been set.
-
ii.Generate a preview slice (optional). A 2D preview stitched image can be generated at a desired z-plane using the preview button.
-
iii.Stitch the dataset under the merge tab. Specify the output directory and the output format.Note: The dataset can also be stitched at downsampled resolutions. A downsampled resolution of the volume is recommended when the dataset is too large and multiple channels need to be combined later in the image processing software such as IMARIS. The TeraStitcher advanced mode can be used to correct misalignment between adjacent tiles. For more information, please refer to TeraStitcher user guide.
-
i.
-
b.The stitched volume can be visualized using IMARIS software (Oxford Instruments).
-
i.Convert the stitch image files into IMARIS format (.ims extension) using IMARIS convertor.Note: You only need to pick the first file from the stitched folder, and the converter automatically recognizes the other files from the consecutive file names.
-
ii.Open the converted file (.ims extension) in IMARIS.
-
iii.Set the associated voxel dimensions in the IMARIS edit/image properties tab.
-
iv.Slice through the 3D volume in IMARIS using the Ortho Slicer tab and adjust the thickness of the plane. Methods video S2 shows an example of stitched 3D volume and the usage of the Ortho Slicer slicing function. The porcine inner ear was stained with anti-MYO7a antibody and imaged with a light-sheet microscope. An example of a stitched volume that was acquired with a confocal microscope is also shown in Methods video S3. In this case, the porcine inner ear was stained with anti-PGP9.5.
-
i.
-
a.
-
2.Basilar membrane tracing (optional)
-
a.In IMARIS, use the “measurement points” tool under the “3D view” tab to assign points manually along the spiral trajectory of either the inner hair cells or the outer hair cells.Note: Set the points along the organ of Corti from apex to base. The spacing between points can be ∼50 μm or less to obtain an accurate tracing.
-
b.Using the save icon under the measurement points tab in IMARIS, save the 3D coordinate of points as a CSV file for further analysis such as the length measurement.
-
a.
-
3.Cellular length measurements along the organ of Corti (optional)
-
a.Measure the cellular length of individual cells using the “measurement points” tool in IMARIS under the “3D view” tab. This tool allows assigning points manually along the length of the cells. The measurement is done under the “slice” view, where you can move between Z-planes accurately and trace the length of individual hair cells that extend across multiple Z-planes.
-
b.Using the save icon under the measurement points tab in IMARIS, save the 3D coordinate of points as a CSV file for further analysis.
-
a.
-
4.Denoising and segmentation (optional but required for cell counting). In this section, extract the areas of interest (e.g., hair cell rows) from the volume. See Methods video S4 and S5 for a step-by-step illustration of denoising and segmentation.
-
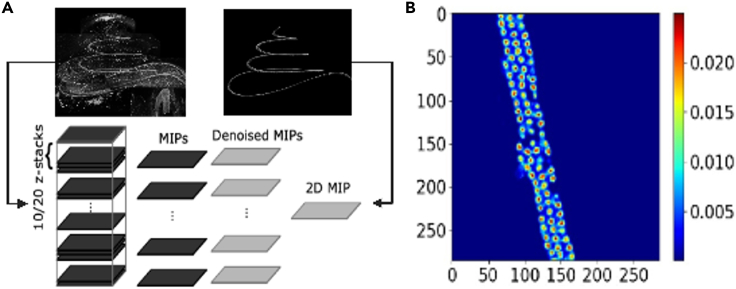
a.Generate partial maximum intensity projection (MIP) for every 10–20 stitched z-planes, using a MATLAB script or ImageJ function (“Z Project” under the “Stack” tab in the “Image” menu). See Figure 6 for a conceptual schematic of the process.
-
b.Denoise the partial MIPs. This will lead to a substantial improvement in the signal-to-noise ratio.
-
i.Use the ImageJ “Selection Brush Tool” to select specific areas of interest such as the hair cells rows in partial MIPs.
-
ii.Use a script or the ImageJ functions (“Make Inverse” function under the “Selection” tab in the “Edit” menu followed by the “Fill” function in the “Edit” menu) to keep only the areas of interest in the partial MIPs.Note: The denoising method removes the background signals of nonspecific antibody binding or autofluorescence outside the regions of interest.
-
i.
-
c.Generate an overall MIP from the denoised partial MIPs using a script or the ImageJ “Z Project” function (Figure 6).
-
d.Segment the areas of interest further to separate cells of interest into inner and outer hair cells using masking in ImageJ (optional). This step will facilitate automated counting.Note: Do step (4d) only if the classes of cells of interest have similar structures such as similar cell body shapes that will make their automatic separation challenging specially if you are using a single antibody staining such as MYO7a.
Methods video S4. Step-by-step illustration of denoising of hair cells, related to step4Download video file (47.4MB, mp4)Methods video S5. Step-by-step illustration of segmentation of hair cells, related to step4Download video file (32.5MB, mp4) -
a.
-
5.Cell counting
-
a.Counting hair cells via ilastik.12 See Methods video S6 for a step-by-step illustration of cell counting. This step is not limited to MIPs of 3D images and can be used for any 2D images.
-
i.Open the ilastik software (version 1.3) and select the Cell Density Counting workflow.
-
ii.Provide the input data, which in our case is the 2D denoised and segmented image that was generated in step 4.Note: Select appropriate features from intensity, edge, and texture based on the pixel size of the cells to be detected by the algorithm. Matching the size of the cells and the scale of selected features is recommended for accurate counting. These features can be changed later if needed.
-
iii.Train the algorithm to detect cells by adding annotation manually via drawing dots on the cell centers of 5–10% of the whole dataset.Note: The ilastik density counting workflow utilizes these annotations to assign labels to pixels based on the pixel features. The “Sigma” feature should roughly match the object size.
-
iv.Train the algorithm to detect the background i.e., regions without cells using brushstrokes.Note: Make sure that the density estimation is close to zero in background regions.
-
v.Use the “live-update” function to obtain the predicted density over the image that was used for training since the training stage is iterative.Note: The training should be done in several regions in the cochlea that varies in their image quality and cellular attributes to generalize well on all the images in the dataset. Train ilastik separately to count different cell types such as inner hair cells and outer hair cells. The features required for accurate counting are different.
-
vi.By placing boxes in specific areas, automated counting results will appear per box. The box can be resized and repositioned.
-
vii.Using the “Current View” menu, switch to another 2D denoised and segmented image that was acquired from a different cochlea. Without annotating the sample, you can either use the “live-update” function to evaluate the model performance or the “Update total intensity” to count all the cells in the entire image.Note: Once the algorithm is well-trained, “batch processing” can be used for the density estimation of similar images.
-
viii.For the quality control, manually count cells in a specific region that is bound by a box and compare that with ilastik automated cells counting results. Do this step at least once per cochlea.Note: If the counting error is more than 5%, retrain ilastik by adding more annotations (sections iii – iv) and repeat. For challenging regions for ilastik, manual counting using ImageJ “Multiple-point Tool” is suggested.
Methods video S6. Step-by-step illustration of cell counting using ilastik, related to step5Download video file (25.6MB, mp4) -
i.
-
a.
Figure 6.
Hair cell counting steps
(A) A schematic of the denoising process, related to step 4.
(B) An image that shows ilastik estimation of cell density, related to step 5.
Limitations
Our proposed tissue clearing and 3D imaging method for studying the inner ear has several inherent limitations such as long processing time per sample, the quenching of endogenous fluorescence (please see Troubleshooting, Problem 4), non-compatibility of all antibodies (please see Troubleshooting, Problem 1), and requirement for the technical expertise of analyzing large datasets of images.
Troubleshooting
Problem 1
Antibody staining, step 3f, did not work as expected.
Potential solution
There could be multiple reasons why the staining has not worked well, we provide a list of solutions:
-
•
Use fresh tissue.
-
•
Increase the primary antibody concentration.
-
•
Use an antibody from a different vendor. Confirm with the vendor that the antibody is compatible with the species.
Note: species-specific antibodies are not available for all biomarkers of interest.
-
•Run a methanol compatibility test, for the desired antibody.13 The test can be performed on any tissue type that robustly expresses the desired protein.
-
○Obtain tissue section ∼ 10-20 μm thick.
-
○Fix the tissue sections with 1% PFA (PBS) for 12–20 h at 4°C (covered)
-
○Wash off PFA three times with PBS.
-
○Incubate tissue sections in 100% methanol for 3 h (covered).
-
○Wash off the methanol three times with PBS.
-
○Permeabilize and block the tissue sections using permeabilization and blocking solutions for 1 h (25°C, covered).
-
○Perform immunostaining with primary antibody for 12–20 h at 4°C.
-
○Wash tissue sections 3× with the washing solution.
-
○Perform immunostaining with secondary antibody for 1 h.
-
○Wash tissue sections 3× with the washing solution.
-
○Mount slides with any mounting media such as ProLong Gold.
-
○Allow slides to dry before imaging.
-
○
Here is the list of antibodies that were tested and found compatible with this method of tissue clearing:
| Antibodies | ||
| Rabbit Anti-CD326 | Sigma | MFCD00212936 |
| Mouse Anti-Acetylated Tubulin | Sigma | MFCD00164512 |
| Mouse anti-CD3 | BioRAD | MCA5951GA |
| Rabbit anti-claudin 4 | Sigma | SAB4200573 |
| Rabbit anti-MYO7a | Proetus | 25-6790 |
| Chicken anti-GFP | Aveslabs | GFP-1020 |
| Rabbit anti-PGP9.5 Polyclonal | Proteintech | 14730-1-AP |
| Rabbit anti-SOX2 | Abcam | 703-605-155 |
| Mouse anti-VGLUT2 | Abcam | ab79157 |
| Rabbit anti-TH | Sigma | AB152, PRID: AB_390204 |
| Rabbit anti-RFP | Rockland | 600-401-379 |
| Rat anti-PECAM1 | bdbiosciences | 553370 |
| Cy™3 AffiniPure Donkey Anti-Rabbit IgG (H + L) | Millipore | 711-165-152 |
| Alexa Fluor® 647 AffiniPure Donkey Anti-Chicken IgY (IgG) (H + L) | Jackson Laboratory | 703-605-155 |
| Alexa Fluor® 647 AffiniPure Donkey Anti-Mouse IgY (IgG) (H + L) | Jackson Laboratory | AB_2340862 |
| Alexa Fluor® 488 AffiniPure Goat Anti-Chicken IgY (IgG) (H + L) | Jackson Laboratory | 103-545-155 |
| Cy3 AffiniPure Donkey Anti-Rat IgY (IgG) (H + L) | Jackson Laboratory | 712-165-153 |
| Nuclear Stain | ||
| TO-PRO-3 | ThermoFisher | T3605 |
Problem 2
The DAPI staining, step 3, did not work.
Potential solution
Use TO-PRO-3 nuclear stain instead which is a red-shifted nuclear stain with excitation at 642 nm.
Note: TO-PRO-3 is very sensitive to bleaching and will fade very quickly. Decrease the laser power and calibrate and focus outside of the area of interest.
Problem 3
The staining, step 3, has worked but the signal looks noisy.
Potential solution
-
•
Use the recommended fixative concentration and timing.
-
•
Use a red-shifted secondary antibody as autofluorescence in the green channel (∼488 nm) is strong.
Problem 4
The endogenous fluorescence is quenched after step 3.
Potential solution
Enhance the signal. For example, in the case of GFP, you can use an anti-GFP antibody.
Problem 5
The sample is not fully transparent after the entire clearing process, step 3.
Potential solution
Change to fresh DBE and check the sample after 12–24 h. You can repeat this step for an additional cycle.
Problem 6
The sample is noisy and ilastik automated counting is not improving in terms of counting accuracy regardless of the amount of manual annotation, step 5 of Quantification and statistical analysis.
Potential solution
The algorithm is overfitting or biased by the current annotation. Restart the training process with more diverse annotation in the training set. Use manual counting in the event that ilastik is not performing well.
Resource availability
Lead contact
Requests for further information about resources and reagents should be directed to the lead contact, Adele Moatti (amoatti@ncsu.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The software programs that were used in this study are listed in the key resources table.
Any additional information required to reanalyze the data reported in this study is available from the lead contact upon reasonable request.
Acknowledgments
The authors would like to thank the NC State University Central Procedure Lab at the College of Veterinary Medicine for their help with tissue collection. This work was supported by funding from the National Institutes of Health (NIH), NIDCD K99 DC019960-01A1, and R21DC020005.
Author contributions
Conceptualization, A.M., A.G., F.S.L.; methodology, A.M., C.L., Y.C., A.G.; investigation, A.M.; visualization, A.M.; resources, K.D.P., K.C.; supervision, A.G., F.S.L.; writing—original draft, A.M.; writing—review & editing, A.M., Y.C., A.G., F.S.L.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2023.102220.
References
- 1.Moatti A., Li C., Sivadanam S., Cai Y., Ranta J., Piedrahita J.A., Cheng A.G., Ligler F.S., Greenbaum A. Ontogeny of cellular organization and LGR5 expression in porcine cochlea revealed using tissue clearing and 3D imaging. iScience. 2022;25:104695. doi: 10.1016/j.isci.2022.104695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Q., Liu K., Yang L., Wang H., Yang J. BoneClear: whole-tissue immunolabeling of the intact mouse bones for 3D imaging of neural anatomy and pathology. Cell Res. 2019;29:870–872. doi: 10.1038/s41422-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutson K.A., Pulver S.H., Ariel P., Naso C., Fitzpatrick D.C. Light sheet microscopy of the gerbil cochlea. J. Comp. Neurol. 2021;529:757–785. doi: 10.1002/cne.24977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urata S., Iida T., Suzuki Y., Lin S.-Y., Mizushima Y., Fujimoto C., Matsumoto Y., Yamasoba T. A novel technique for imaging and analysis of hair cells in the organ of Corti using modified Sca/eS and machine learning. Bio. Protoc. 2019;9 doi: 10.21769/BioProtoc.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moatti A., Cai Y., Li C., Sattler T., Edwards L., Piedrahita J., Ligler F.S., Greenbaum A. Three-dimensional imaging of intact porcine cochlea using tissue clearing and custom-built light-sheet microscopy. Biomed. Opt Express. 2020;11:6181–6196. doi: 10.1364/BOE.402991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rai M.R., Li C., Greenbaum A. Quantitative analysis of illumination and detection corrections in adaptive light sheet fluorescence microscopy. Biomed. Opt Express. 2022;13:2960–2974. doi: 10.1364/BOE.454561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li C., Moatti A., Zhang X., Troy Ghashghaei H., Greenabum A. Deep learning-based autofocus method enhances image quality in light-sheet fluorescence microscopy. Biomed. Opt Express. 2021;12:5214–5226. doi: 10.1364/BOE.427099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bria A., Iannello G. TeraStitcher - a tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. BMC Bioinf. 2012;13:316. doi: 10.1186/1471-2105-13-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ollion J., Cochennec J., Loll F., Escudé C., Boudier T. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics. 2013;29:1840–1841. doi: 10.1093/bioinformatics/btt276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arshadi C., Günther U., Eddison M., Harrington K.I.S., Ferreira T.A. SNT: a unifying toolbox for quantification of neuronal anatomy. Nat. Methods. 2021;18:374–377. doi: 10.1038/s41592-021-01105-7. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira T.A., Blackman A.V., Oyrer J., Jayabal S., Chung A.J., Watt A.J., Sjöström P.J., van Meyel D.J. Neuronal morphometry directly from bitmap images. Nat. Methods. 2014;11:982–984. doi: 10.1038/nmeth.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg S., Kutra D., Kroeger T., Straehle C.N., Kausler B.X., Haubold C., Schiegg M., Ales J., Beier T., Rudy M., et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods. 2019;16:1226–1232. doi: 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 13.Renier N., Wu Z., Simon D.J., Yang J., Ariel P., Tessier-Lavigne M. iDISCO: a Simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Three sensory organs are shown: the saccule, posterior crista, and cochlea, related to step 4.
4
4
5
Data Availability Statement
The software programs that were used in this study are listed in the key resources table.
Any additional information required to reanalyze the data reported in this study is available from the lead contact upon reasonable request.