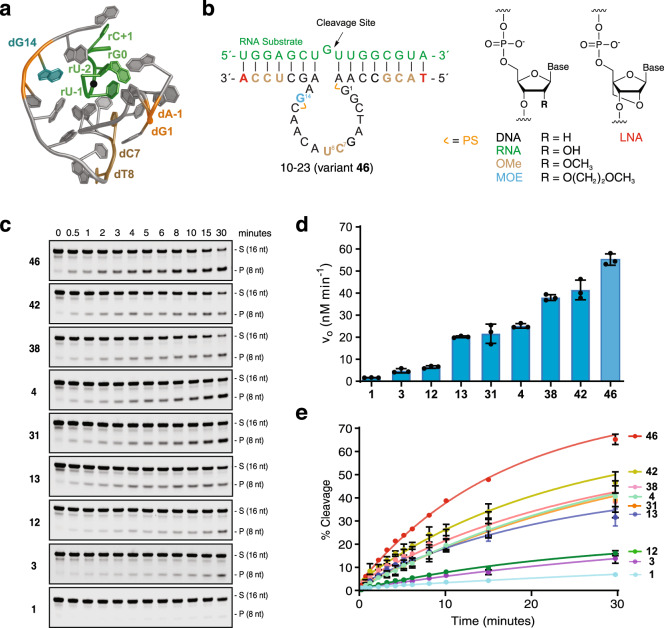
Fig. 1. Chemical evolution of DNAzyme 10-23.
a Cartoon representation of an NMR-averaged precatalytic structure of 10–23 bound to an RNA substrate (green) encoding a prion protein (“7PDU”). Dz is shown in gray, with colors and numbers matching residues found to be important in the chemically optimized version. b Left: Dz 10-23_v46 in complex with a 16-nt KRAS G12V RNA substrate. Right: Chemical structures of natural and modified nucleotides found in Dz 10-23_v46. Abbreviations: DNA (2′-deoxyribose nucleic acid), RNA (ribose nucleic acid), OMe (2′-deoxy-2′-methoxyribonucleic acid), MOE (2′-deoxy-2′-methoxyethoxyribonucleic acid), LNA (locked nucleic acid) and PS (phosphorothioate). Modifications are colored to denote their position in Dz 10-23. c Representative denaturing PAGE gels showing time-dependent multiple turnover RNA cleavage activity across a panel of engineered Dz 10–23 variants. S: 5’-Cy5-labeled full-length substrate, P: 5′-Cy5-labeled cleavage product. d, e Initial velocities (vo, D) and kinetic curves (E) were measured for each variant. Error bars denote the ±standard deviation of the mean for three independent replicates. All reactions were performed under simulated physiological conditions in a buffer containing 1 mM MgCl2, 50 mM Tris (pH 7.5), 10 mM NaCl, and 140 mM KCl at 37 °C with 1000 nM substrate and 10 nM enzyme (100:1, S:E). Source data are provided as a Source Data file.