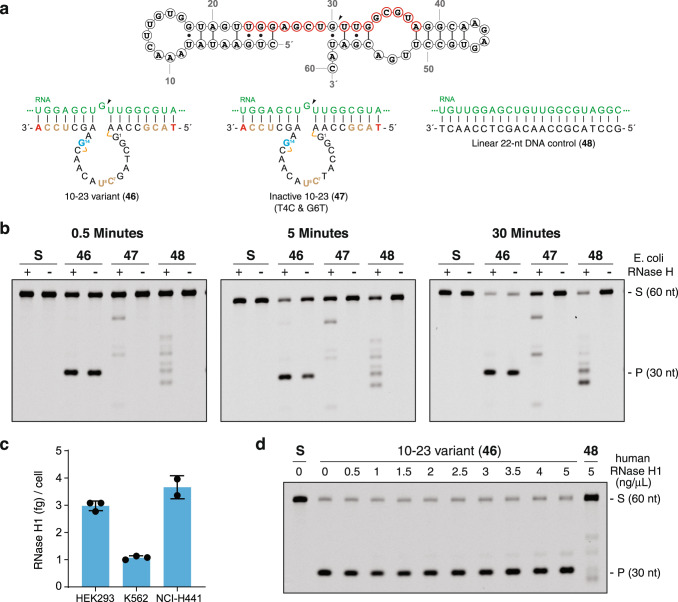
Fig. 3. Autonomous DNAzyme-mediated RNA cleavage mechanism.
a Reagents used to evaluate the mechanism of RNA cleavage include a 60 nt RNA segment of KRAS G12V, Dz 10-23_v46 (active Dz), Dz 10–23_v47 (inactive Dz), and a 22 nt linear DNA. The DNAzyme recognition site on the RNA substrate is shown in red, and the RNA cleavage site is indicated with an arrow. The color scheme for DNAzyme-substrate pairs is the same as shown in Fig. 1b. b Representative denaturing PAGE gels showing RNA cleavage profiles in the presence (+) or absence (−) of E. coli RNase HI (0.05 U/μL) after 0.5, 5, and 30 min of incubation (Dz 46 and 47, n = 3; 48, n = 1). c Quantification of RNase H1 in three mammalian cell lines by ELISA. Cellular levels of RNAse H1 range from 0.5 to 2.5 ng/μL. Error bars denote the ±standard deviation of the mean for three independent replicates. d Denaturing PAGE gel showing RNA cleavage profiles in the absence (−) or presence (+) of human RNase H1 after a 30 min incubation (n = 1). Linear DNA is a positive control. Reactions were performed in a buffer containing 1 mM MgCl2, 50 mM Tris (pH 7.5), 10 mM NaCl, and 140 mM KCl at 37 °C with 250 nM substrate and 250 nM enzyme (1:1, S:E). S: 5′-Cy5-labeled full-length substrate, P: 5′-Cy5-labeled cleavage product. Source data are provided as a Source Data file.