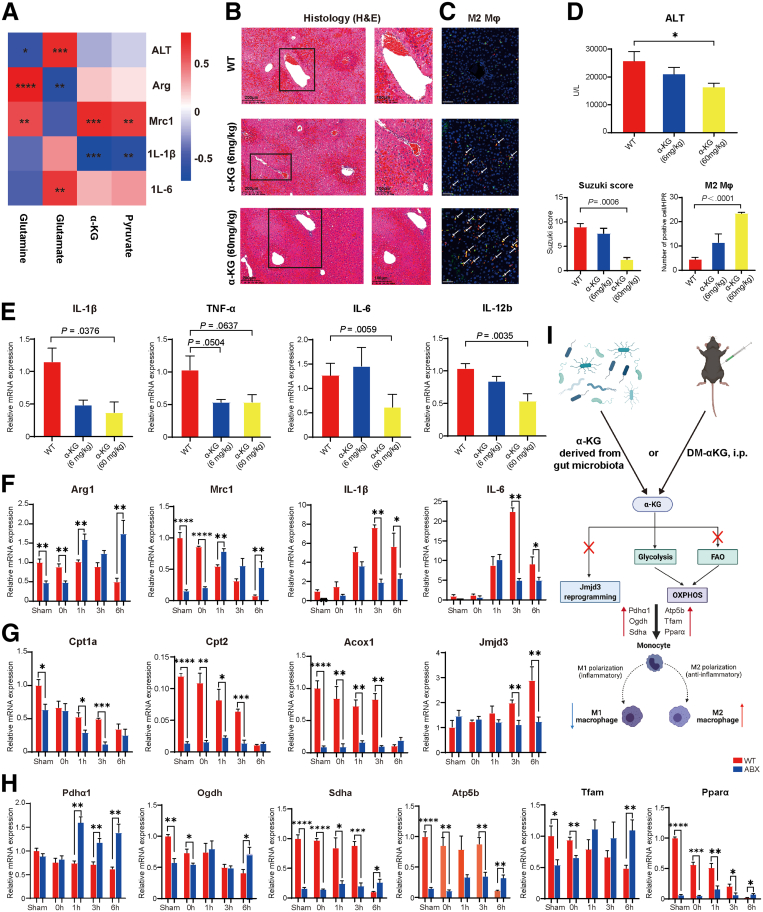
Figure 7.
Serum αKG could lead to macrophage metabolic reprogramming to promote M2 phenotype differentiation via the OXPHOS metabolic pathway. (A) Heatmap of the correlation analysis between the fecal levels of glutamine, serum glutamate, αKG, and pyruvate and the disease indicator ALT as well as the expression of the M1 or M2 gene markers Il1β, Il6, Arg1, and Mrc1. (B) H&E staining and Suzuki’s quantitative score of the WT, αKG (6 mg/kg, n = 5), and αKG (60 mg/kg, n = 5) groups. (C) Immunofluorescence double labeling of M2 macrophages and semiquantitative analysis. Scale bars, 100 μm. (D) Serum ALT levels of the 3 groups. (E) Relative mRNA expression of the M1 marker genes Il1β, Tnfα, Il6, and Il12b in the 3 groups. (F) Relative mRNA expression of the M2 marker genes Arg1 and Mrc1 and the M1 marker genes Il1β and Il6 at different time points. (G) Relative mRNA expression of the FAO metabolic pathway-related genes Cpt1a and Cpt2 and the epigenetic gene Jmjd3. (H) Relative mRNA expression of the OXPHOS metabolic pathway-related genes Pdhα1, Ogdh, Sdhα, Atp5b, Tfam, and Pparα. The polymerase chain reaction data above show the kinetics of macrophage M1/M2 marker expression, n = 3–4 for each group. (I) Mechanistic diagram of how αKG can protect the liver from I/R injury. For all data, statistical comparisons between 2 groups were carried out by Student t test. Correlation comparison was performed by Spearman’s correlation analysis. P < .05 indicates significant differences.