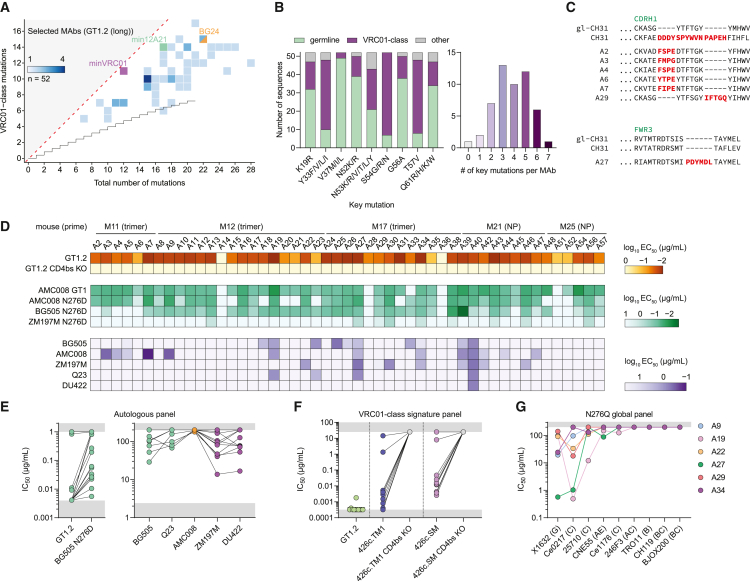
Figure 4.
GT1.2-primed VRC01-class mAbs display diverse binding and neutralization capacities
(A) Total and VRC01-class amino acid mutations in the IGHV1-2 region as in Figure 3D for the 52 selected mAbs, including minVRC01 (purple), min12A21 (green), and BG24 (orange).
(B) Stacked bar graph (left) showing whether the residue at the positions indicated on the x axis are germline (green) has mutated into a key VRC01-class residue (purple) or another residue (gray). (Right) Distribution of the number of key mutations in each individual mAb.
(C) Amino acid alignment showing multi-residue insertions (red) in the CDRH1 (top) or in the FWR3 (bottom) of selected mAbs.
(D) Midpoint binding titers (EC50) of each of the selected mAbs. The color corresponds to the starting concentration used (orange, 1 μg/mL; green, 10 μg/mL; purple, 50 μg/mL).
(E) Midpoint neutralization titers (IC50) of a subset of selected mAbs. Each dot corresponds with an individual mAb.
(F) Midpoint neutralization titers (IC50) of a subset of selected mAbs against VRC01-class signature viruses.
(G) Midpoint neutralization titers (IC50) of a subset of selected mAbs against an N276Q global panel as indicated on the x axis.