Figure 5.
Rare insertions and deletions are important for mAb-Env interactions and might establish quaternary contacts
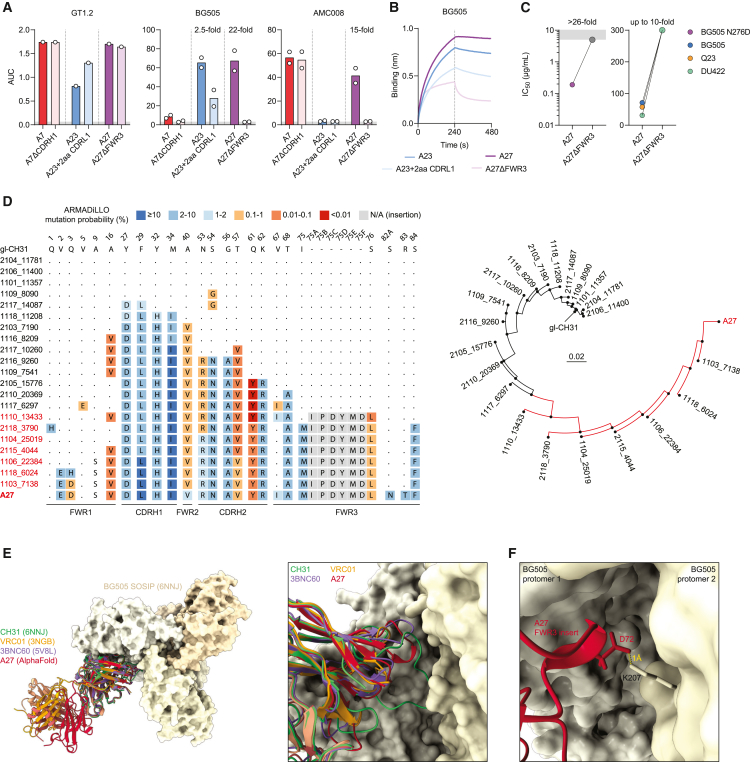
(A) ELISA binding to Envs expressed as the area under the curve (AUC) for each of the original mAbs and mAbs that had their specific indel removed. Each dot represents an individual experiment.
(B) Bio-layer interferometry (BLI) sensorgrams showing binding of A23 (blue) and A27 (purple) with their indel events removed.
(C) Midpoint neutralization titers (IC50) of A27 and A27ΔFWR3 against the viruses indicated.
(D) Amino acid sequence alignment with ARMADiLLO mutation probabilities (left) and phylogenetic tree (right) of an A27 lineage reconstructed by identifying the shortest path between gl-CH31 and A27 using NGS repertoire reads from the mouse from which A27 was isolated.
(E) Structural representation (top view) of a BG505 SOSIP Env (PDB: 6NNJ) in complex with bNAbs CH31 (PDB: 6NNJ), VRC01 (PDB: 3NGB), 3BNC60 (PDB: 5VBL), and an AlphaFold2-Multimer-modeled structure of A27.
(F) Side view of residue D72A27 in the FWR3 insertion extending toward K207gp120.