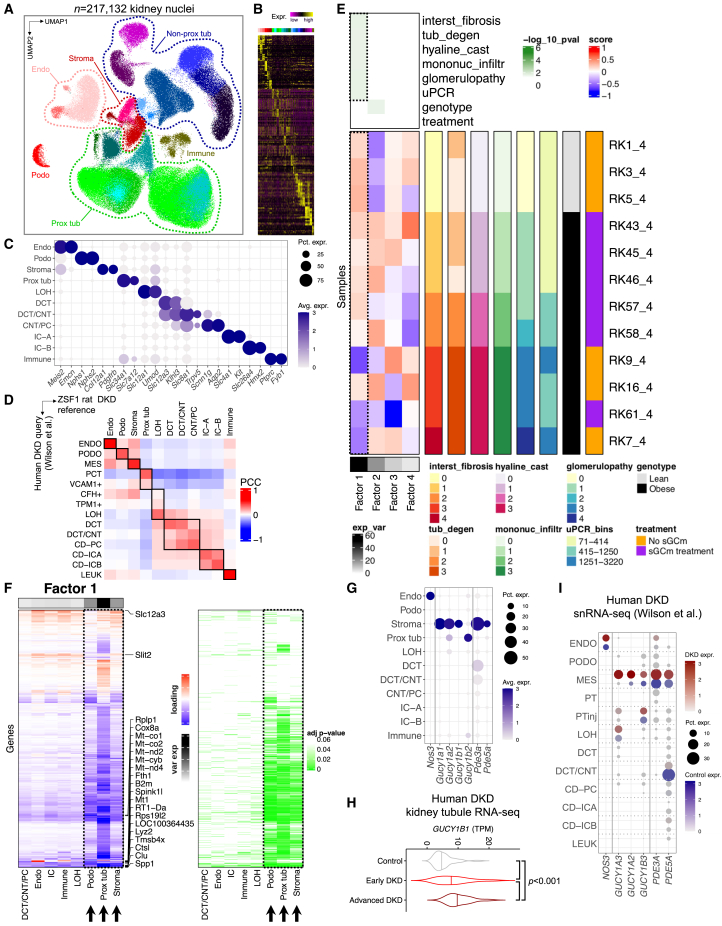
Figure 2.
Single-cell transcriptomic landscape of the diabetic ZSF1 rat
(A) Integrated UMAP of 217,132 high-quality nuclei from twelve rat kidney samples; Endo, endothelial cells; Immune, immune cells; Non-prox tub, non-proximal tubule; Podo, podocytes; Prox tub, proximal tubule; Stroma, stromal cells.
(B) Heatmap of top ten differentially expressed genes for low-level clustering.
(C) Marker gene expression for high-level clustering. Dot size denotes percentage of cells expressing the marker. Color scale represents average gene-expression values.
(D) Pearson correlation coefficient (PCC) matrix of average cell type gene expression between ZSF1 rats (lean and obese samples only) and a corresponding human snRNA-seq dataset with control and DKD kidney samples. CD-ICA/IC-A, collecting duct intercalated cell type A; CD-ICB/IC-B, collecting duct intercalated cell type B; CD-PC/PC, collecting duct principal cell; CNT, connecting tubule; DCT, distal convoluted tubule; Endo/ENDO, endothelial cell; LEUK, leukocyte; LOH, loop of Henle; MES, mesangial cells; PCT, proximal convoluted tubule; Podo/PODO, podocytes; Prox tub, proximal tubule.
(E) Tensor decomposition analysis heatmap (center left) representing factor loading score of rat kidney (RK) samples (rows) onto tensor factors (rows). Degree of explained variance (exp_var) in the whole dataset is displayed on the bottom left. Significance level (−log10(p value)) of tensor factor association with clinical (uPCR, urinary protein/creatinine ratio in mg/mmol) and histopathology outcome measures (interst_fibrosis, interstitial fibrosis; tub_degen, tubular degeneration; mononuc_infiltr, mononuclear infiltration, glomerulopathy, each scored from 0 to 4) is displayed on the top left. Sample rows are color-annotated by outcome data, genotype (lean vs. obese), and treatment status (sGCm, sGC modulator treatment, or no treatment).
(F) Heatmap representing factor 1 loading scores by cell type (columns) and genes (rows) (left). Explained variance is colored in shades of gray (top left), significance levels are shown on the right. The top five significant genes for every cell cluster are annotated.
(G) Expression dot plot for NO/sGC/cGAMP pathway genes. Dot size denotes percentage of cells expressing the marker. Color scale represents average gene-expression values.
(H) Expression of GUCY1B1 in human microdissected kidney tubule bulk RNA-seq samples, stratified by control, early DKD, and advanced DKD cases; p value is given for one-way ANOVA (Tukey corrected). TPM, transcripts per million.
(I) Expression dot plot for NO/sGC/cGAMP pathway genes in a human DKD snRNA-seq dataset. Dot size denotes percentage of cells expressing the marker. Red and blue color scales represent average gene-expression values in DKD and control samples, respectively; PTinj, injured PT (composite of VCAM1+, CFH+, TPM1+ PT cells).