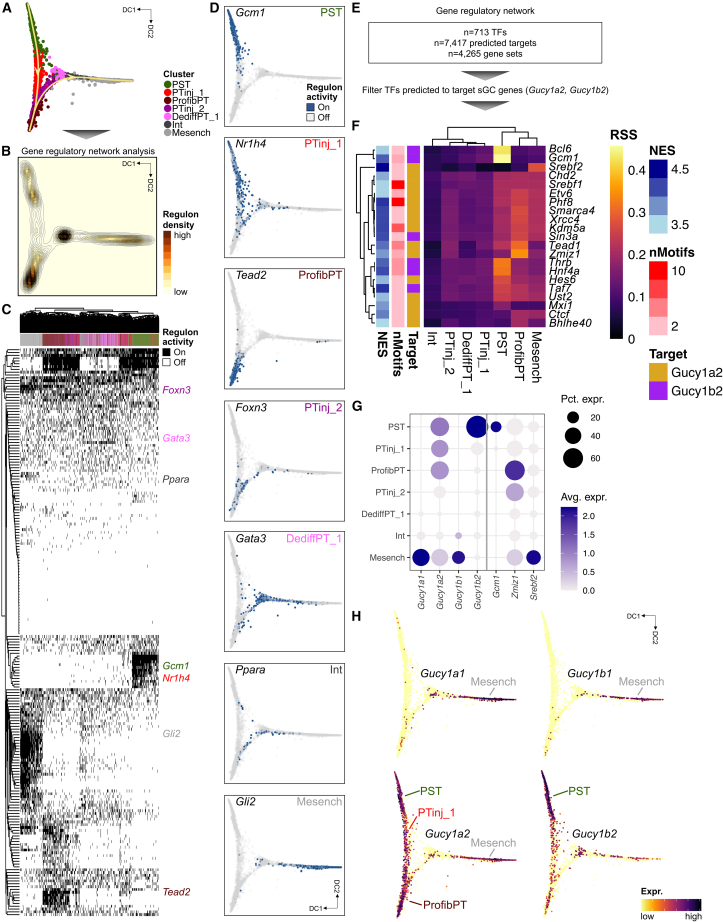
Figure 6.
Gene-regulatory network analysis highlights cell-type-specific transcription factors driving the PT-to-mesenchymal trajectory and prioritizes cell types of action for sGC modulation
(A) Cell clusters from the PT-Mesench trajectory were subjected to gene-regulatory network (GRN) analysis.
(B) Regulon density as a surrogate for stability of regulon states along the trajectory in diffusion map space.
(C) Heatmap of cell-type-specific binarized regulon activity. Rows represent regulons of transcription factors (TFs) and their predicted targets, columns represent cells along the trajectory, colored by cell clusters as in (A). Top specific TFs per cluster are annotated.
(D) Binarized regulon activity for top cluster-specific TFs along the trajectory in diffusion map space.
(E) The GRN dataset was filtered for TFs predicted to target sGC genes (Gucy1a2, Gucy1b2).
(F) Heatmap visualizing specificity of regulons (rows) for cell clusters along the trajectory (columns). Color denotes regulon specificity score (RSS). Regulons are color-annotated for normalized enrichment score (NES), number of motifs, and their predicted sGC target gene (Gucy1a2 or Gucy1b2). Top cell-cluster-specific regulons are annotated.
(G) Expression dot plot for sGC genes and top cell-cluster-specific TFs from (F). Dot size denotes percentage of cells expressing the marker. Color scale represents average gene-expression values.
(H) Feature plots for sGC genes (Gucy1a1, Gucy1a2, Gucy1b1, Gucy1b2) along the trajectory in diffusion map space.