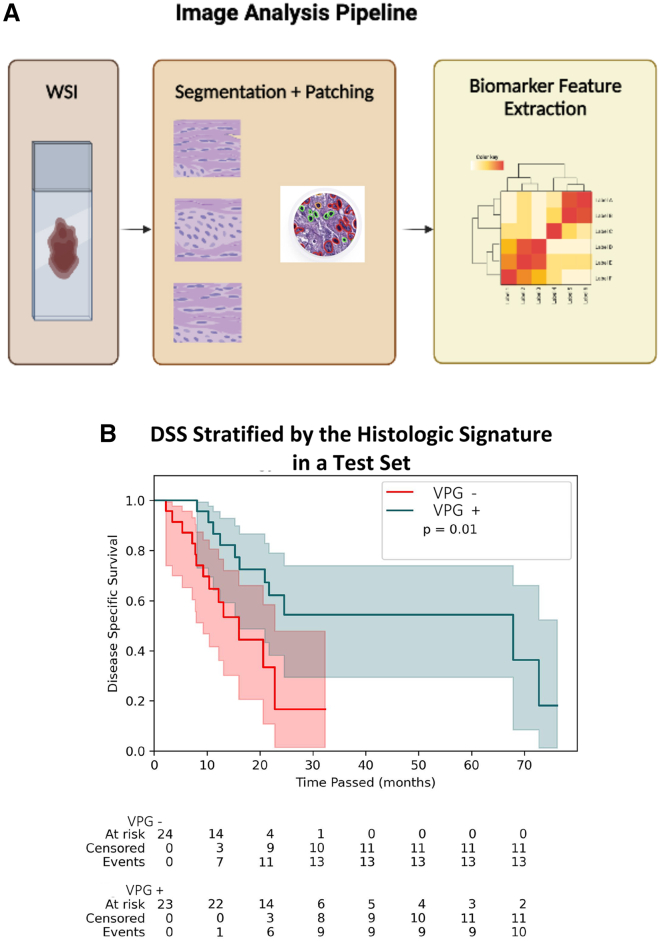
Figure 2.
An image analysis pipeline yields a histologic signature that stratifies disease-specific survival (DSS) following adjuvant gemcitabine
(A) In the image analysis pipeline for this study, whole-slide images (WSIs) from tumor resections were converted to smaller patches before cell-level segmentation and geometric feature extraction describing cellular morphology at the patient level. 816 features are extracted for each individual patient from the available digitized slide. Downstream statistical analysis of these features enables identification of a histologic signature.
(B) Kaplan-Meier curves for the test set from TCGA cohort (n = 47) stratified by the presence or absence of the AI-derived histologic signature. The p value (p = 0.01) corresponds to the log rank test. The median DSS for signature+ patients was 67.9 months (95% CI: [16.2, not reached]), and the median DSS for signature− patients was 16 months (95% CI: [9.3, 22.8]).