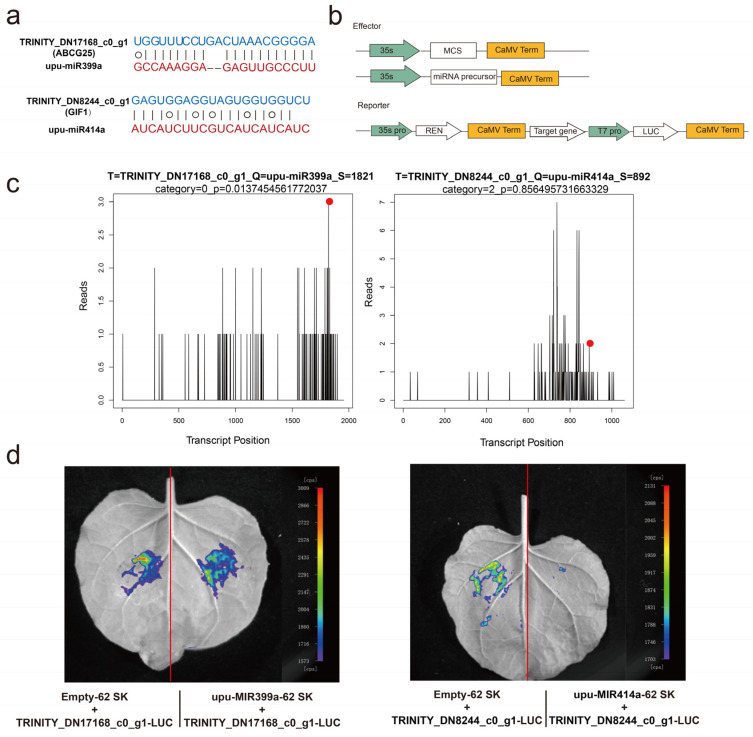
Figure 6.
Validation of miRNA cleavage predicted target genes by dual−luciferase assay. (a) Sequence match between upu−miR399a, upu−miR414a, and their predicted targets. The target gene (top) and its corresponding miRNA (bottom) are shown in each alignment. The match is displayed in a joint line, and G: U wobble pairing in a circle. (b) Construction of the reporter and effector vectors of a dual−luciferase assay. (c) Target plots (t−plots) of miRNA targets confirmed by degradome sequencing. The vertical line with a red point indicates the cleavage site of each transcript. (d) Dual−luciferase assay of upu−miR399a−ABCG25 (left panel) and upu−miR414a−GIF (right panel). The left side of the red line is the injection area of the control group, and the right side is the injection area of the experimental group. Different colors mean luciferase activity. Cps, signal counts per second.