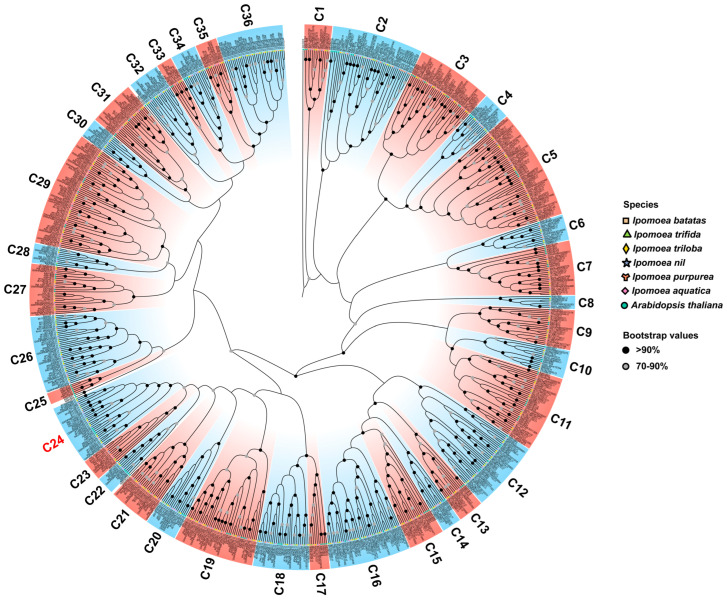
Figure 2.
Phylogenetic tree of R2R3-MYB members in Arabidopsis and six Ipomoea species. All identified R2R3-MYB members from I. batatas (131), I. trifida (133), I. triloba (129), I. nil (127), I. purpurea (51), I. aquatica (124), and Arabidopsis (126) were used. The full-length amino acid sequences of R2R3-MYB proteins were aligned using MAFFT, and the tree was constructed by the maximum-likelihood (ML) method of IQ-tree using the JTT + F + R10 model. These R2R3-MYB proteins are clustered into 36 clades (designated as C1 to C36). The AtCDC5 was rooted as the outgroup, and 4 proteins did not fit well into the clade. The genes that belonged to the same organism were marked in the same shape and color. The dots on the branches represent bootstrap values based on 1000 replications, and bootstrap values >90% and ≥70% are shown as black and gray dots in the phylogenetic tree, respectively, while those <70% are not shown.