Table 4.
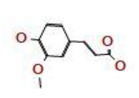
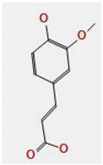
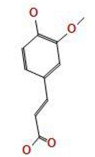
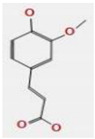
Docking scores and energies of ferulic acid with crystal structure of H. pylori 4HI0.
| Mol (Five Poses of Ferulic Acid) | rseq | mseq | S | rmsd_refine | E_conf | E_place | E_score1 | E_refine | E_score2 |
|---|---|---|---|---|---|---|---|---|---|
|
1 | 1 | −5.58 | 3.35 | −42.86 | −61.32 | −11.22 | −27.30 | −5.58 |
|
1 | 1 | −5.35 | 1.81 | −40.17 | −64.29 | −11.15 | −29.20 | −5.35 |
|
1 | 1 | −5.31 | 2.06 | −41.95 | −63.97 | −9.95 | −28.10 | −5.31 |
|
1 | 1 | −5.29 | 1.73 | −41.47 | −67.91 | −10.93 | −27.96 | −5.29 |
|
1 | 1 | −5.24 | 2.00 | −40.25 | −74.10 | −9.96 | −28.98 | −5.24 |
S, the final score (score of the last stage that was not set to none); rmsd, the root-mean-square deviation of the pose, in Å, from the original ligand; rmsd_refine, the root-mean-square deviation among the pose before refinement and the pose after refinement; E_conf, the energy of the conformer; E_place, the score from the placement stage; E_score 1 and E_score 2, the score from rescoring stages 1 and 2, respectively; E_refine, the score from the refinement stage, which is determined to be the totality of the solvation energies and van der Waals electrostatics, according to the generalized Born solvation model.
