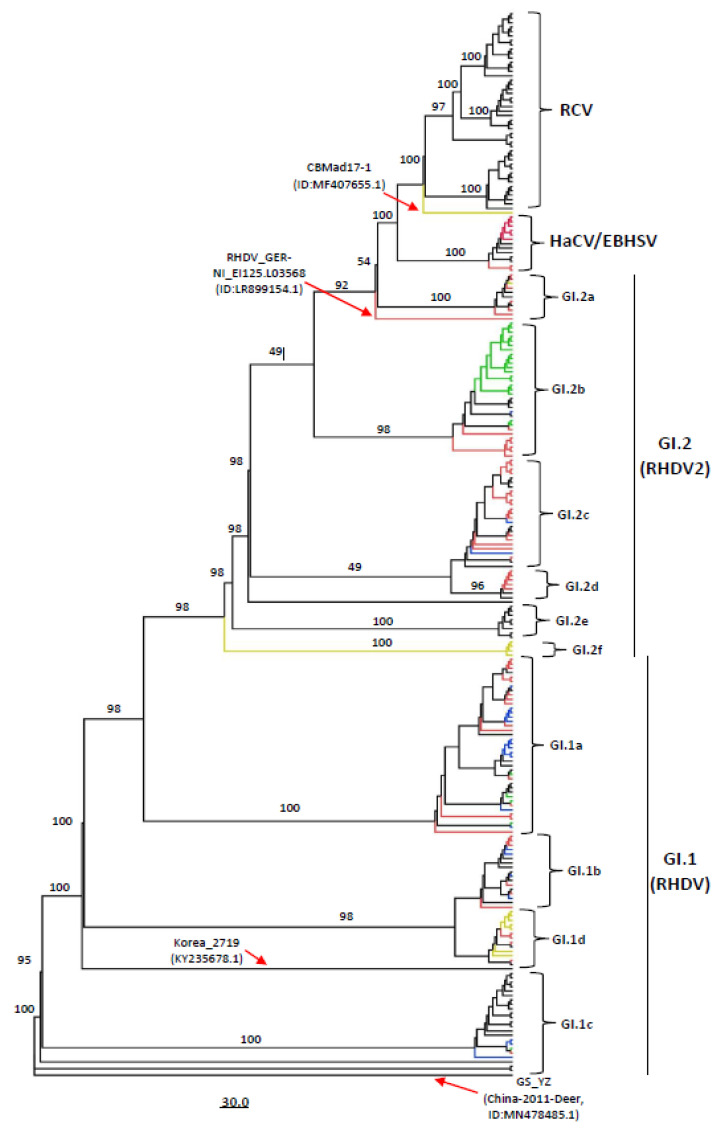
Figure 1.
The phylogenetic tree based on the full-length genome sequences of Lagovirus strains, 1988–2021. The unrooted ML phylogenetic tree of 240 full-length Lagovirus genomes classifying all the strains into four major clades (GI.1/RHDV, GI.2/RHDV2, RCV, and HaCV/EBHSV). GI.1 can be further classified into four sub-clades (GI1a–d) and RHDV2/GI.2 into six sub-clades (GI.2a–f). The major clades and sub-clades of the RHDV are indicated. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are indicated at each node. The evolutionary distances were computed using the best-fit substitution model (SYM + I + G 4). The red color clades represent Germany (n = 69), green USA (n = 24), blue Poland (n = 22), and the yellow color represents Portugal (n = 13). The tree was visualized and modified to proportion using FigTree v1.4.