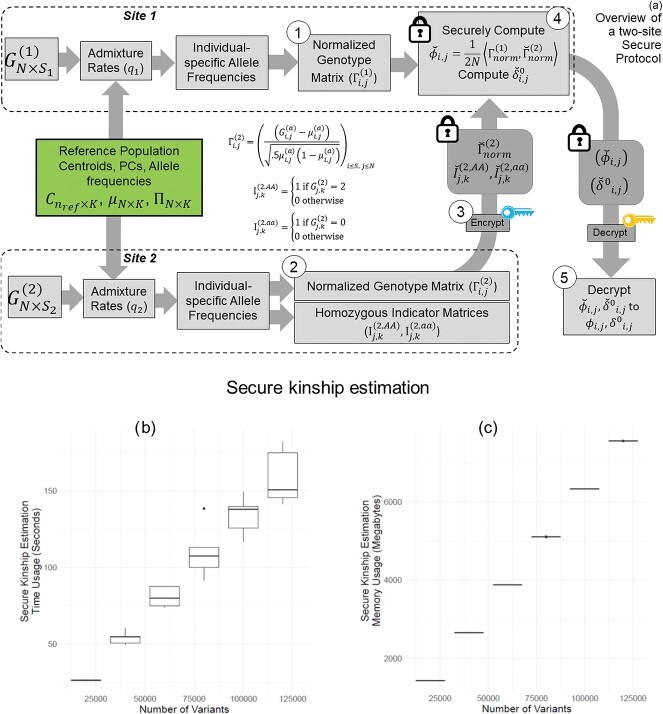
Figure 6.
Illustration of secure kinship and IBD-Sharing probability estimation for 2-site collaboration. (A) Site-2 estimates individual specific allele frequencies and computes the normalized genotypes 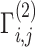 , and indicator matrices
, and indicator matrices 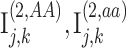 , and sends them to Site-1 after encrypting them with the public key. Site-1 also computes the normalized genotype matrix,
, and sends them to Site-1 after encrypting them with the public key. Site-1 also computes the normalized genotype matrix, 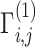 and the indicator matrices. After receiving the encrypted genotype matrix from Site-2, Site-1 securely estimates the encrypted kinship (
and the indicator matrices. After receiving the encrypted genotype matrix from Site-2, Site-1 securely estimates the encrypted kinship (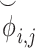 ) and zero-IBD sharing probability matrix (
) and zero-IBD sharing probability matrix (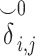 ) shown in step 4. Site-1 sends the encrypted matrices to Site-2, which decrypts the kinship statistics and shares them with Site-2. (B) The time requirements of the secure kinship estimation (y-axis in seconds) of the HAPMAP project’s 86 MEX samples with respect to increasing number of variants (x-axis). (C) Memory requirements of secure kinship estimation (y-axis in megabytes) with respect to increasing number of variants (x-axis).
) shown in step 4. Site-1 sends the encrypted matrices to Site-2, which decrypts the kinship statistics and shares them with Site-2. (B) The time requirements of the secure kinship estimation (y-axis in seconds) of the HAPMAP project’s 86 MEX samples with respect to increasing number of variants (x-axis). (C) Memory requirements of secure kinship estimation (y-axis in megabytes) with respect to increasing number of variants (x-axis).