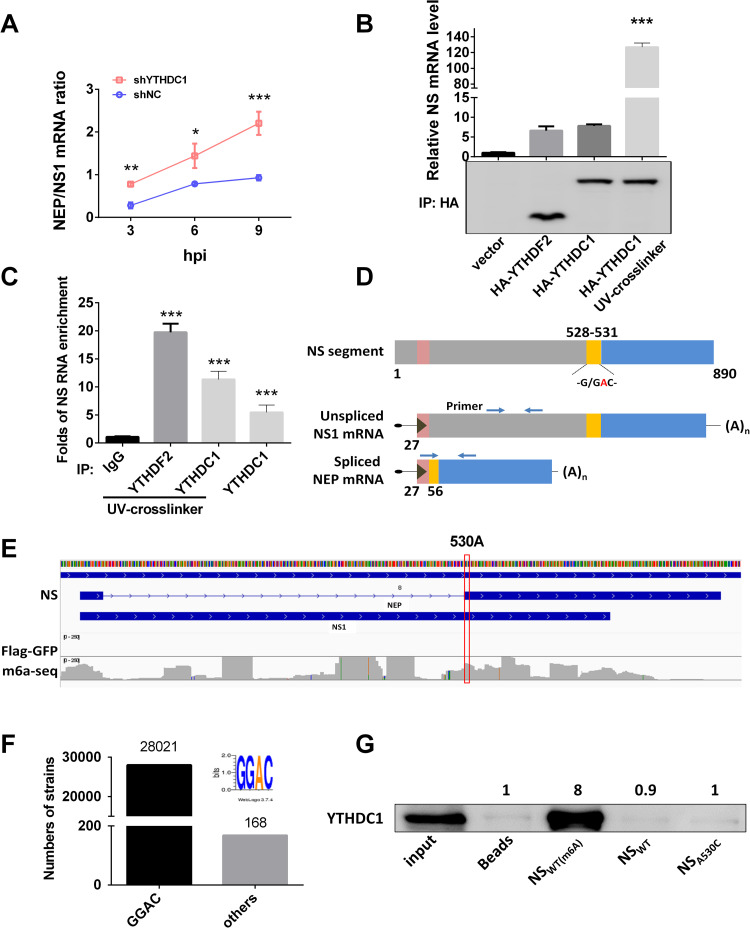
Fig 3. NS1 inhibits NS segment splicing by YTHDC1.
(A) YTHDC1 knockdown (shYTHDC1) or negative control (shNC) knockdown A549 cells were infected with the PR8 virus at an MOI of 5. The ratios of NEP to NS1 mRNA were calculated at indicated time points with the specific primers. Data are presented as the average of three experiments and error bars indicate the standard error of the mean (SEM) (two-way ANOVA test; * P<0.05, ** P<0.01; ***, P<0.001). (B) HEK293T cells were transfected with indicated plasmids for 24 h and then infected with the PR8 virus at an MOI of 1 for 12 h. RIPs and RT-qPCR were performed using specific primers detecting viral NS mRNA. Fold enrichment of mRNA was calculated. Data are presented as the average of three experiments and error bars indicate the standard error of the mean (SEM) (one-way ANOVA; ***, P<0.001). (C) A549 cells were infected with the PR8 virus at an MOI of 1 for 12 h. RIPs and RT-qPCR were performed using specific primers detecting viral NS mRNA. Fold enrichment of mRNA was calculated. Data are presented as the average of three experiments and error bars indicate the standard error of the mean (SEM) (one-way ANOVA; ***, P<0.001). (D) NS1 mRNA and its alternatively spliced product were depicted, and the arrowheads show primer positions for detection of various. G/GAC (528–531) is the YTHDC1-binding site and ‘/’ is the 3′ splicing site in the NS segment. (E) A view of the NS segment of IAV-PR8. PA-m6A-seq has a y-axis of 0–250 reads[14]. The 530A site is marked in the red box. (F) Analysis of GGAC motif in NS 528–531 segment of IAVs, total of 28189 strains were analyzed. (G) Pulldown YTHDC1 proteins using 20 bp NS biotin-probes of PR8: probes were immunoprecipitated with the cell lysis. And the bead eluate was analyzed by western blotting.