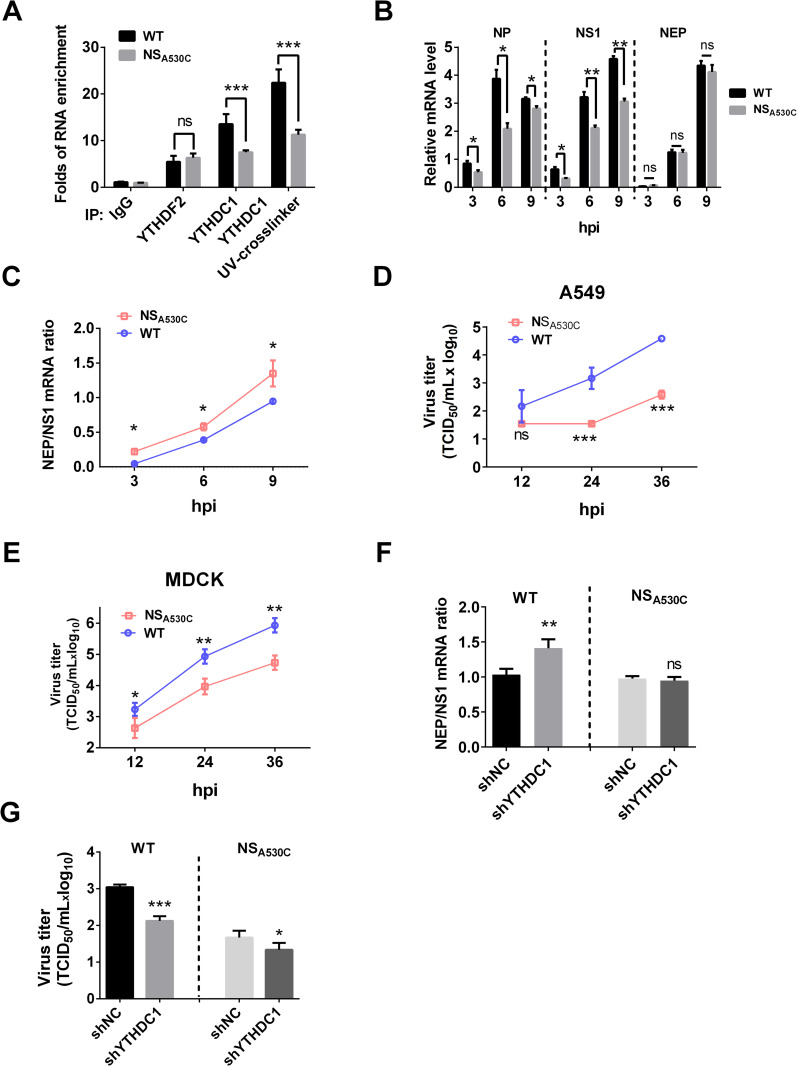
Fig 5. NSA530C mutation altered virus splicing and replication.
(A)A549 cells were infected with the wild type or mutated PR8 virus at an MOI of 1 for 12 h, and the mRNAs were purified with either the control IgG or the YTHDC1 antibody. The purified RNAs were then analyzed by specific RT-qPCR assay. Data are presented as the average of three experiments and error bars indicate the standard error of the mean (SEM) (one-way ANOVA; ns, not significant; ***, P<0.001). (B) A549 cells were infected with either the wild type or mutant of the PR8 virus at an MOI 5 for the indicated periods. The total RNA was isolated and analyzed by specific RT-qPCR targeting NP, NEP and NS1 mRNA, which normalized to GAPDH mRNA. Data are presented as the average of three experiments and error bars indicate the standard error of the mean (SEM) (two-way ANOVA; ns, not significant; *, P<0.05; **, P<0.01; ***, P<0.001). (C) A549 cells were infected with the indicated virus at an MOI of 5, the total RNA was isolated and analyzed by specific RT-qPCR targeting NEP and NS1 mRNA. Ratios of NEP/NS1 mRNA are presented. Data are presented as the average of three experiments and error bars indicate the standard error of the mean (SEM) (two-way ANOVA; *, P<0.05). (D-E) (D)A549 cells and (E) MDCK cells were infected with either the wild type or mutant of the PR8 virus at an MOI 0.01 for the indicated periods. The growth curves were determined by TCID50 analysis on MDCK cells. Data are presented as the average of three experiments and error bars indicate the standard error of the mean (SEM) (two-way ANOVA; ns, not significant; **, P<0.01; ***, P<0.001). (F) A549 cells were infected with the wild type (WT) and mutant (NSA530C) PR8 virus at an MOI of 5. The total RNAs were measured by RT-qPCR and the NEP/NS1 mRNA ratios were calculated. Data are presented as the average of three experiments and error bars indicate the standard error of the mean (SEM) (one-way ANOVA; ns, not significant; **, P<0.01; ***, P<0.001). (G) A549 cells were infected with the wild type (WT) and mutant (NSA530C) PR8 virus at an MOI of 0.01. The virus titers were detected at 24 hpi. Data are presented as the average of three experiments and error bars indicate the standard error of the mean (SEM) (one-way ANOVA; ns, not significant; **, P<0.01; ***, P<0.001).