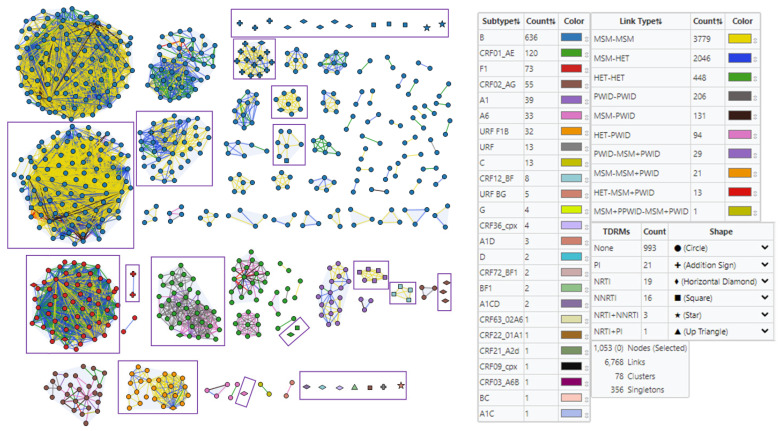
Figure 2.
Identification and characterization of HIV transmission networks in antiretroviral-naive Bulgarians using MicrobeTrace. HIV-1 polymerase (pol) sequence clusters identified using a genetic distance cutoff (d) of 1.5%. Nodes are colored by final subtype classification using a combination of sequence analysis methods. Node shape indicates type of resistance mutation: PI, protease inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitor. Clusters are grouped by size and subtype, and singletons with surveillance drug resistance mutations (SDRMs) are also shown, whereas those non-clustering sequences without resistance mutations were excluded. Clusters and singletons with SDRMs are shown in boxes. Edges (links) are colored by transmission risk category (MMSC, male-to-male sexual contact; HET, heterosexual transmission; PWID, people who inject drugs; MMSC + PWID, MMSC who also report injecting drugs).