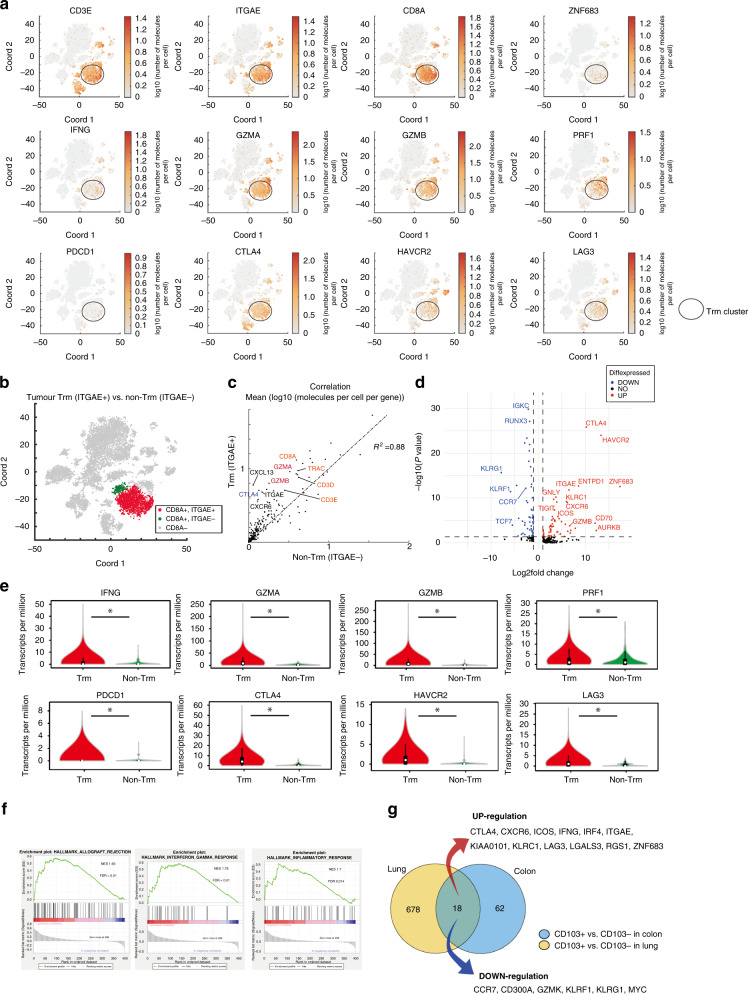
Fig. 2. Trm cells represent the most activated subset of CD8+ T cells in CRC.
a t-SNE plots of ~12,000 live and singlet-gated CD45+ single-cell transcriptomes obtained from resected CRC tissue showing gene expression encoding Trm cell markers, cytotoxic cytokines, and immune checkpoint molecules. b Trm (red) and non-Trm (green) clusters are defined according to ITGAE (encoding CD103) and CD8A expression. c Correlation between Trm and non-Trm cell clusters: mean (log10 [molecules/cell/gene]). d Volcano plot showing DEGs between Trm and non-Trm cell clusters. Dot lines represent an FDR of <0.05 and a fold change of >2. e Violin plot of the expression of cytotoxic cytokines and immune checkpoint molecule genes in the two clusters in (b). f Top 3 NES of hallmark gene sets in GSEA between Trm and non-Trm cell clusters. g Venn diagrams overlapping transcripts differentially expressed with a fold change of >2 between Trm and non-Trm cell clusters in lung cancer (left) and CRC (right). Trm cells tissue-resident memory T cells, CRC colorectal cancer, ITGAE integrin subunit alpha E, t-SNE t-distributed stochastic neighbour embedding, DEG differentially expressed gene, FDR false discovery rate, NES normalised-enrichment scores, GSEA gene set enrichment analysis.