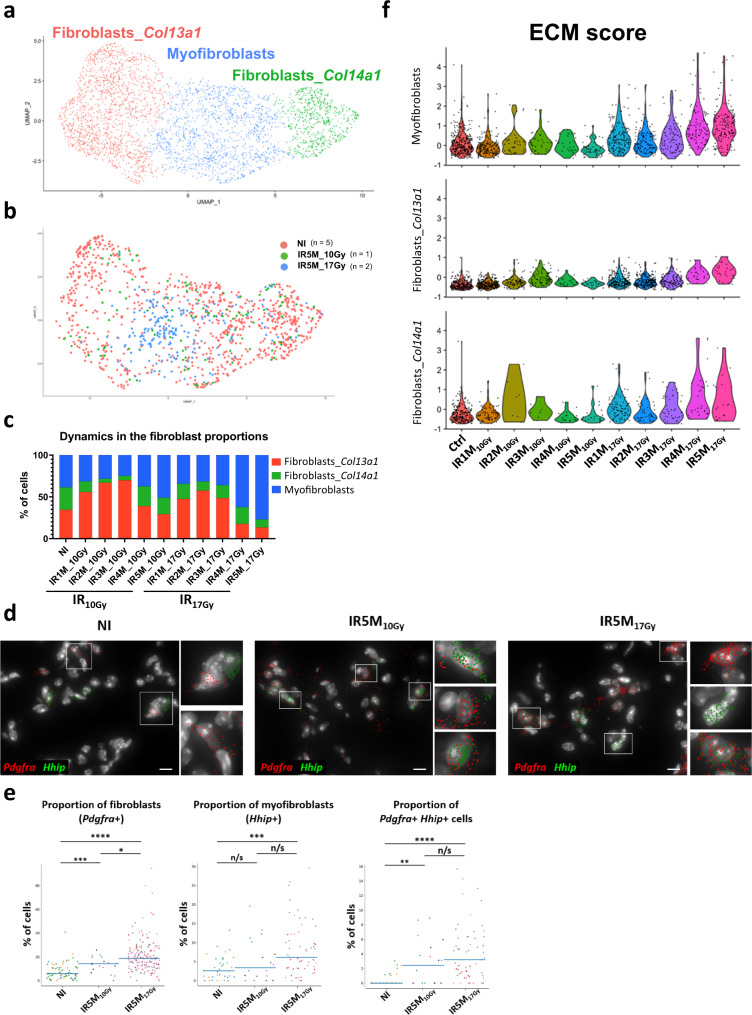
Fig. 3. Myofibroblasts contribute to the ECM deposition after IR17Gy.
a UMAP visualization of 3488 cells from the different fibroblast subpopulations annotated by cell type. b UMAP visualization of NI (n = 5), IR5M10Gy (n = 1) and IR5M17Gy (n = 2) fibroblasts annotated by time point. c Dynamics in the proportion of the fibroblast subpopulations at the different time points after IR10Gy and IR17Gy. d Automatic Pdgfra (red) and Hhip (green) mRNA detection with Big-FISH in NI, IR5M10Gy, and IR5M17Gy lung tissue sections. Scale bars, 10 µm. e Quantification of the Pdgfra+, Hhip+ and Pdgfra+ Hhip+ cells in the NI, IR5M10Gy and IR5M17Gy lung tissue sections. To compare two groups, the P value was computed with the Mann–Whitney–Wilcoxon test (two-sided test) from scipy (n/s, adjusted p value >0.05; *, adjusted p value <0.05; **, adjusted p value <0.01; ***, adjusted p value <0.001; ****, adjusted p value <0.0001). Each dot represents one analyzed image. Each color per time point represents a different biological replicate (NI n = 3; IR5M10Gy n = 3; IR5M17Gy n = 5). f Violin plot showing the single cell score calculated based on the ECM expressed genes in the myofibroblasts, fibroblasts Col13a1, and fibroblasts Col14a1.