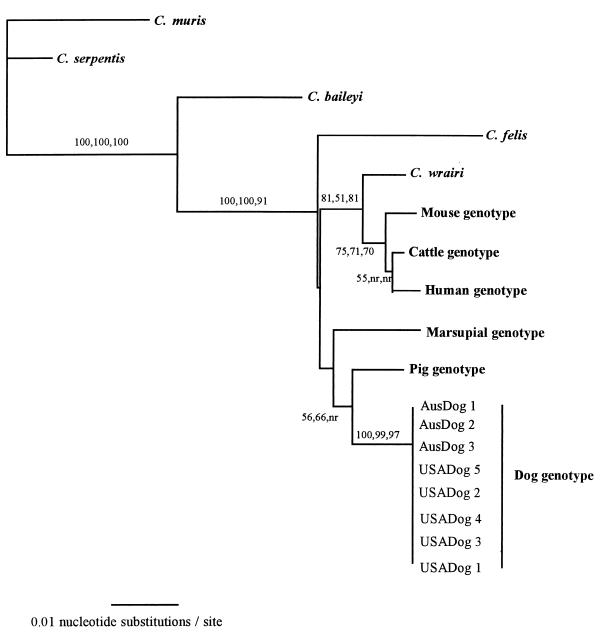
FIG. 2.
Evolutionary relationships of Cryptosporidium isolates inferred by NJ analysis of Tamura-Nei distances calculated from pairwise comparisons of 18S rDNA sequences. Percentages of bootstrap support (>50%) from 1,000 replicate samples (analyzed by the NJ, parsimony, and maximum-likelihood methods, respectively) are indicated at the left of the supported node. nr, node not recovered by method. Additional Cryptosporidium 18S rDNA sequences obtained from GenBank were bovine C. parvum isolate C1 (AF108864), human C. parvum isolate H7 (AF108865), mouse genotype C. parvum isolate M24 (AF108863), a pig genotype isolate (AF108861), a marsupial genotype isolate (AF108860), C. wrairi (AF115378), C. felis (AF108862), C. baileyi (AF093495), and C. serpentis (AF108866).