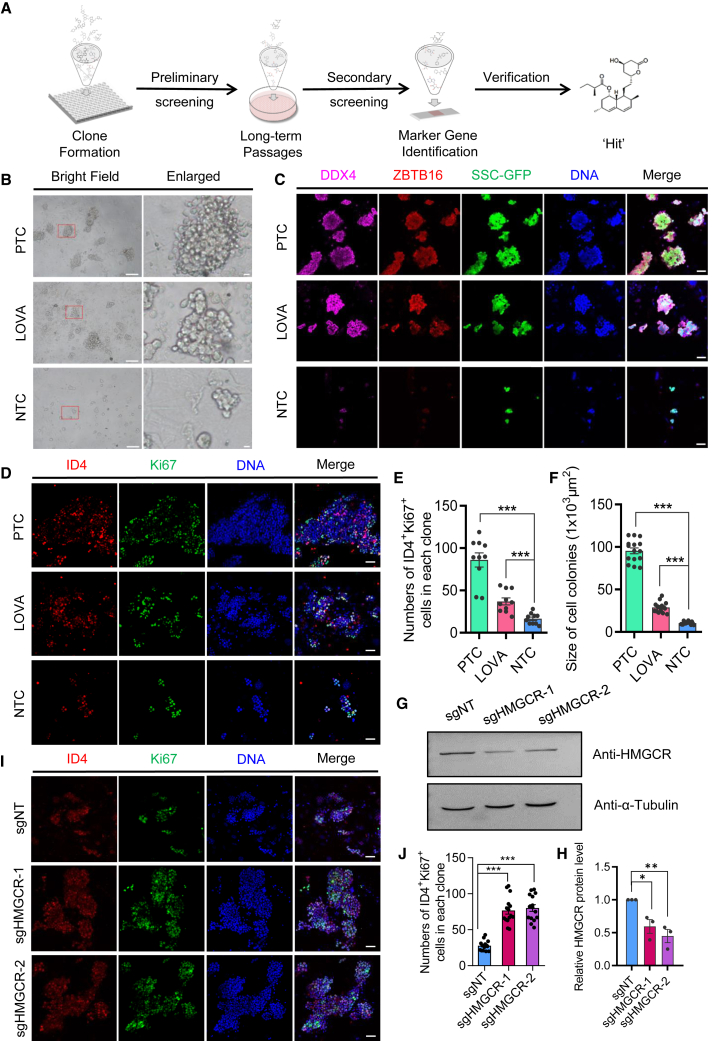
Figure 1.
Lovastatin maintains the identity of mSSCs in vitro in the absence of GDNF
(A) Schematic outline of the small-molecule screening.
(B) Morphology of mSSC clones in different culture conditions. mSSCs cultured in regular conditions that contain exogenous GDNF were named the positive control group (PTC). mSSCs cultured without GDNF were used as the negative control group (NTC). Small-molecule compound-treated mSSC group (Lovastatin [LOVA]) was cultured in NTC medium supplemented with LOVA. The right panel shows the clone enlarged in the red rectangle of left field. Scale bars: left, 100 μm, and right, 10 μm.
(C) Immunofluorescent staining of mSSC clones cultured in different groups for 18 days. GFP indicates mSSC clones seeded in the culture dish. DDX4 and ZBTB16 as germ cell and undifferentiated spermatogonia markers were labeled, respectively. Scale bars, 50 μm.
(D) Immunofluorescent staining of mSSC clones cultured in different groups for 18 days. ID4 and Ki67 as mSSCs and cell proliferation markers were labeled, respectively. Scale bars, 50 μm.
(E) Statistic histogram of ID4 and Ki67 double-positive cells for each group from ten independent experiments. Error bars indicate mean ± SEM, ∗∗∗p ≤ 0.001.
(F) Statistic histogram of size of mSSC clones for each group from fifteen independent experiments. Error bars indicate mean ± SEM, ∗∗∗p ≤ 0.001.
(G) Western blot of HMGCR knockdown mediated by CRISPR-Cas13d system. sgHMGCR-1 and sgHMGCR-2 were designed to knock down HMGCR, while sgNT indicates non-targeting sgRNA. α-Tubulin was used as the loading control.
(H) Statistic histogram of relative HMGCR protein level in (G) from three independent experiments. Error bars indicate mean ± SEM, ∗p ≤ 0.05, ∗∗p ≤ 0.01.
(I) Immunofluorescent staining of HMGCR knockdown and control sgNT mSSCs clones used in (G). mSSC samples were cultured withouth GDNF for 3 passages (18 days) after HMGCR or sgNT knockdown. ID4 and Ki67 as mSSCs and cell proliferation markers were labeled, respectively. Scale bars, 50 μm.
(J) Statistic histogram of ID4 and Ki67 double-positive cells for each group in (I) from ten independent experiments. Error bars indicate mean ± SEM, ∗∗∗p ≤ 0.001.
See also Figure S1.