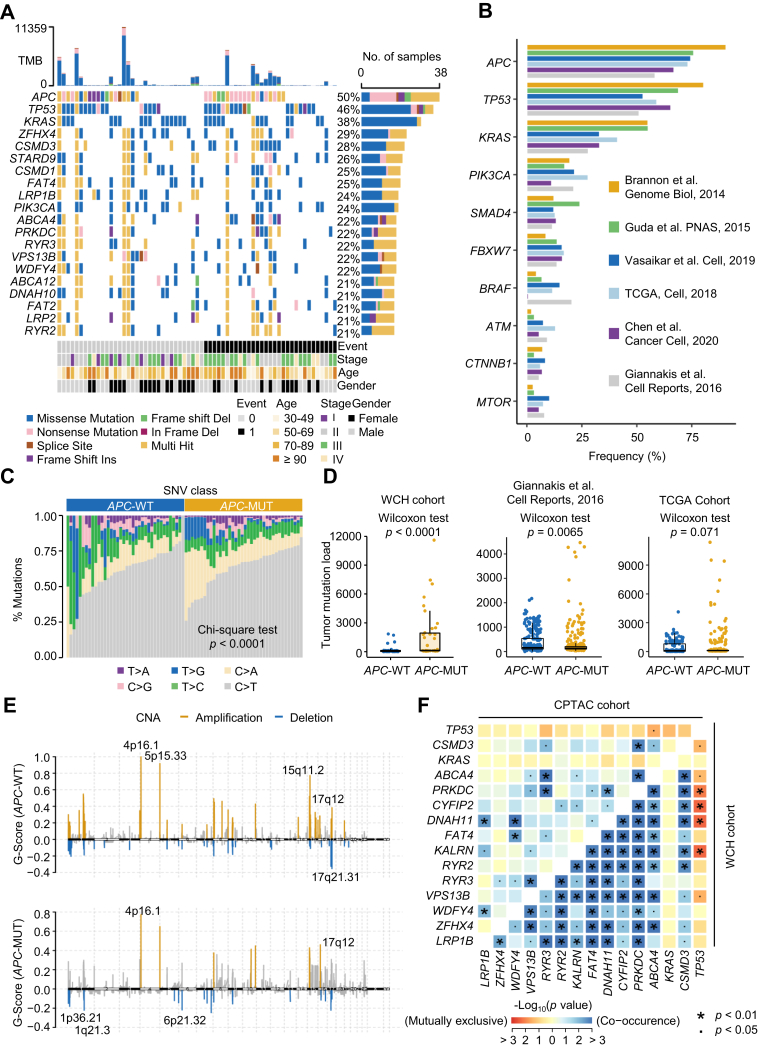
Fig. 1.
Genomic comparisons between APC-MUT and APC-WT colon cancer.A, genetic profile and associated clinicopathologic features of all 76 colon cancer patients. The bar plot on the top indicates the total number of somatic mutations in each patient. The bar plot on the right represents the distribution and compositions of mutation types in each gene. B, bar plot of mutational frequency (%) for the top ten mutated genes in the previously published CRC databases. C, percentages of SNVs in APC-WT and APC-MUT tumors. Test methods, Chi-square test. D, comparison of tumor mutation load between APC-MUT and APC-WT tumors in the WCH cohort, Giannakis’ study, and TCGA cohort. Test methods, Wilcoxon rank-sum test. E, focal peaks with significant somatic copy number amplification (yellow) and deletion (blue) (GISTIC2 Q-values <0.1) shown in APC-MUT and APC-WT tumors, respectively. The top five amplified and deleted cytobands are labeled. F, the relationship between the somatic mutation frequencies of the first 15 mutated genes in APC-MUT tumors of the WCH cohort (bottom right), and the relationship of somatic mutation frequencies of the same 15 genes (top left) in APC-MUT tumors of the CPTAC cohort. Test methods, Fisher’s exact test. APC, Adenomatous polyposis coli; APC-MUT, APC-mutant; CRC, colorectal cancer; CPTAC, Clinical Proteomic Tumor Analysis Consortium; GISTIC, Genomic Identification of Significant Targets in Cancer; SNV, single nucleotide variant; TMB, Tumor mutation burden; WCH, West China Hospital.