Figure 1. Human GU-motif removal and MS2-directed tethering demonstrate TDP-43 binding locus, while cryptic site mutations identify TDP-43 dependent misprocessing requiring cryptic splice acceptor.
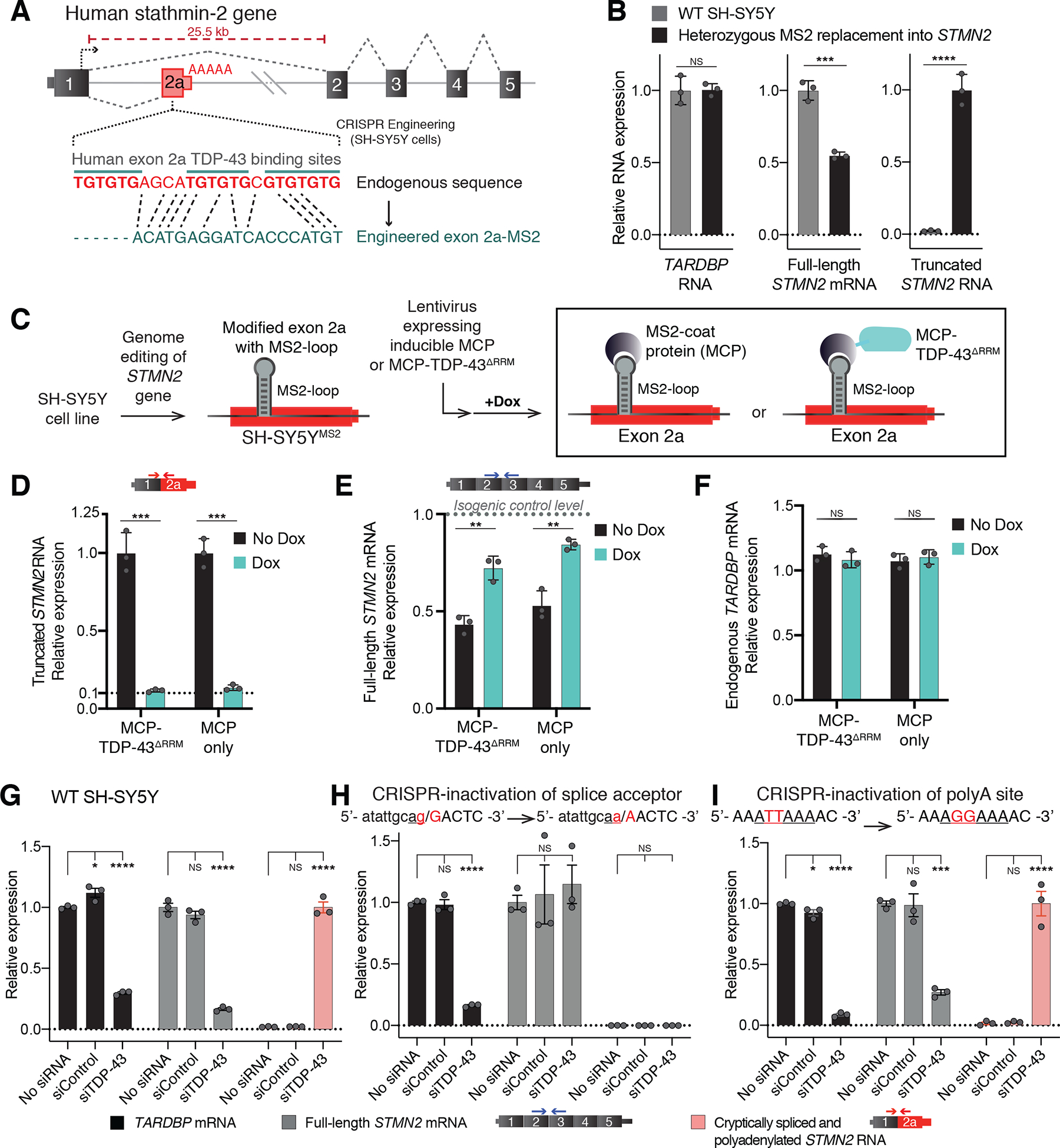
(A) Schematic of CRISPR-engineering strategy for conversion of the GU binding motif in exon 2a into an MS2 aptamer sequence in one STMN2 allele of diploid SH-SY5Y neuroblastoma cells. (B) qRT-PCR demonstrating that SH-SY5Y cells carrying heterozygous GU to MS2 edit misprocess STMN2 RNA, leading to 50% loss of stathmin-2 encoding mRNAs compared with wildtype cells and accumulation of truncated RNA. (C) Schematic depicting MS2:MCP directed strategy to direct MCP-tethered proteins to the normal TDP-43 binding locus. (D-F) qRT-PCR measurement of (D) truncated STMN2, (E) full-length STMN2 mRNA, or (F) endogenous TARDBP mRNA levels with and without induction of MCP-fusion protein expression in SH-SY5Y cells carrying heterozygous MS2 aptamer insertion. (G-I) qRT-PCR measured expression of TARDBP, full-length STMN2 mRNA and truncated STMN2 RNAs 96 hours after siRNA treatment with a control siRNA pool or a pool targeting TARDBP in (G) wildtype SH-SY5Y cells, (H) SH-SY5Y cells harboring homozygous mutation of the human exon 2a 3’ splice acceptor site and (I) SH-SY5Y cells harboring a homozygous mutation of the human exon 2a premature polyadenylation signal to the murine sequence AGGAAA. For all qPCR analysis individual data points are independently treated wells of cells. Error bars are SEM. Statistical significance was determined by 2-tailed Student’s T-test (B-F), or 1-way ANOVA with Dunnett correction (G-I). ****, p <0.0001; ***, p < 0.001; **, p < 0.01; *, p <0.05.
