Abstract
Background/Aim: Epithelial ovarian cancer (EOC) is usually diagnosed in advanced stages and has a high mortality rate. In this study, we used the proximity extension assay from Olink Proteomics to search for new plasma protein biomarkers to predict overall survival (OS) in patients with EOC.
Materials and Methods: Peripheral blood samples were obtained preoperatively from 116 EOC patients undergoing primary debulking surgery: 28 early EOC cases (FIGO stage I-II) and 88 advanced EOC cases (FIGO stage III-IV). Proteins were measured using the Olink Oncology II and Inflammation panels. In total, 177 unique protein biomarkers were analysed. Cross-validation and LASSO regression were combined to select prediction models for OS.
Results: The model including age and the three-biomarker combination of neurotrophin-3 (NT-3)+transmembrane glycoprotein NMB (GPNMB)+mesothelin (MSLN) predicted worse OS with AUC=0.79 (p=0.004). Adding cancer antigen 125 (CA125) and human epididymis protein 4 (HE4) to the model further improved performance (AUC=0.83; p=0.003). In a postoperative model including age and stage (III+IV vs. I+II), the three-biomarker panel of chemokine (C-C motif) ligand 28 (CCL28)+T-cell leukaemia/lymphoma protein 1A (TCL1A)+GPNMB improved the prediction of OS (from AUC=0.83 to AUC=0.90; p=0.05). In the postoperative model including age and dichotomized stage (III vs. I+II), the biomarkers CCL28 and GPNMB1 improved the prediction of OS (AUC=0.86; p<0.001). The combination of high levels of both CA125 and HE4 predicted worse survival (p=0.05).
Conclusion: In this explorative study evaluating the performance of plasma protein biomarkers in predicting OS, we found that adding biomarkers, especially NT-3, to the panel improved the prediction of OS.
Keywords: Epithelial ovarian cancer, overall survival, biomarker
Epithelial ovarian cancer (EOC) is considered a silent carcinoma because it is usually diagnosed in advanced stages. Patients with borderline ovarian tumours have an excellent prognosis, but the prognosis is poor for patients with advanced EOC. Despite advances in ovarian cancer therapy, half of patients will die within five years (1,2). Various biomarkers and their combinations have been tested for the detection of EOC. CA125 has commonly been used since the early 1990s, but it has limitations. A biomarker panel consisting of HE4, CA125 and age can improve discrimination between malignant and benign ovarian tumours (3). HE4 has been found to be an independent marker for shorter progression-free survival and shorter overall survival (4,5). However, a gold standard has not been found. In the exploration for new biomarker panels, we chose to use the Olink® Oncology II and Inflammation panels, which are broad and well-established protein panels that use very small amounts of plasma.
A series of studies has shown that immunological components play a key role in cancer development. Ovarian cancer can create a complex tumour microenvironment with ascites consisting of a mixture of various immunosuppressive cells that impairs the ability of the patient’s immune system to fight the disease (6). A variety of cytokines, chemokines and growth factors are present in EOC (7-9). The aim of the study was to search for new protein biomarkers and biomarker panels.
Materials and Methods
Patients and samples. Peripheral blood samples were obtained preoperatively from 180 women with an adnexal mass admitted for surgery at the Department of Obstetrics and Gynecology, Skåne University Hospital Lund, Sweden, from 2005 to 2012. Blood was collected in citrate tubes and centrifuged, and the plasma was stored at −20˚C until analysed. All diagnoses were verified by histopathological examination. The disease was staged, and morphology was analysed according to the International Federation of Gynecology and Obstetrics (FIGO). No cancer patient had received neoadjuvant chemotherapy. The patient cohort included 28 early EOC cases (FIGO stage I-II) and 88 advanced EOC cases (FIGO stage III-IV). The frozen plasma samples were sent to Olink Proteomics AB, Uppsala, Sweden, for analyses.
Proximity extension assay. Proteins were measured using the Olink Oncology II and Inflammation panels (Olink Proteomics AB, Uppsala, Sweden) according to the manufacturer’s instructions. Included biomarkers in each panel have been listed in Leandersson et al. (10). Proximity extension assay (PEA) technology has been described previously (11). The analyses were performed at Olink Proteomics AB in Uppsala, Sweden. The technicians performing the analyses were unaware of the patient disease status. Samples were randomized on the plates and run-in duplicates. Data were quality controlled and normalized using an internal extension control as well as an interplate control to adjust for intra- and internal variation. All assay validation data are available on the manufacturer’s website.
Six EOC cases did not pass internal quality control in the PEA analyses and were excluded from statistical analyses due to either large intracorrelation variance or inability to read one of the duplicate samples.
Statistical analyses. To investigate whether combinations of proteins were associated with survival at 60 months, a combination of cross-validation and LASSO regression was employed. Overall survival at 60 months was chosen since many studies report 5-year survival. We split the data randomly into a training set and a test set. The shrinkage parameter (λ) was estimated using k-fold cross-validation in the training set. To perform variable selection, the estimated shrinkage parameter λCV was then used in the test set. The selected variables and the absolute value of the coefficients were saved, and the process was repeated 10 times. The variables were then ordered by the number of times they were selected and the sum of their estimated coefficients. The lowest ranked variable was removed, and the entire process was repeated until a final model was selected. This method has also been described by Leandersson et al. (2020) (10). The final models were estimated with logistic regression and predicted probabilities from this model were used to assess the model’s discriminatory abilities. Receiver operator curves (ROCs) were constructed, and the area under the curve (AUC) was calculated with 95% confidence intervals using the nonparametric bootstrap procedure. Cross-validation and LASSO regression analyses were carried out using R v 4.1.0 (12).
Availability of data and materials. All data were obtained according to the Swedish Act concerning the Ethical Review of Research Involving Humans to ensure confidentiality and are available on reasonable request.
Ethics statement. Written informed consent was obtained from all study participants. Ethical approval was granted by the Ethical Review Board at the Faculty of Medicine, Lund University, Sweden. Dnr 495 2016 (amendment to Dnr 558–2004 and 94–2006).
Results
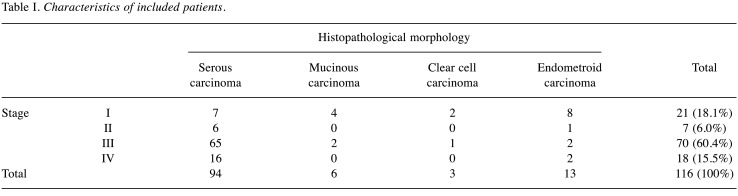
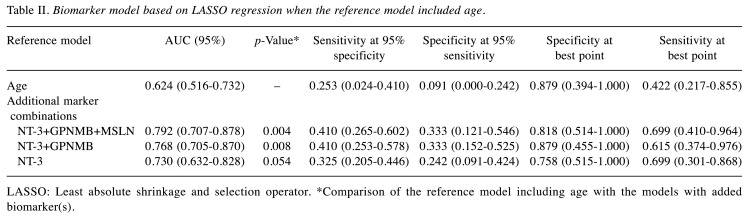
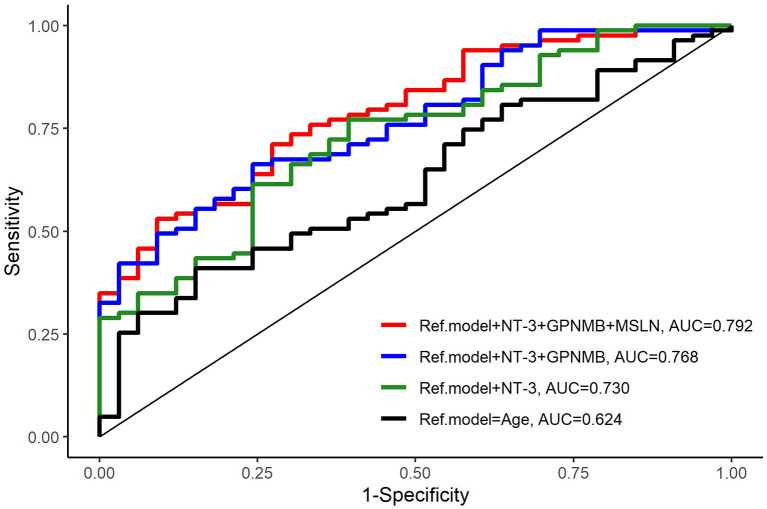
Plasma from 116 EOC patients with different histopathological morphologies was included in the analyses (Table I). The diagnostic performance of a reference model including age only [AUC=0.624 (0.516-0.732)] for discrimination of survival was poor. The biomarker NT-3 alone and in combination with GPNMB and MSLN improved the diagnostic performance, with the best results for the model including age and the three-biomarker panel combination of NT-3+GPNMB+MSLN [AUC=0.792 (0.707-0.878); p=0.004] (Table II, Figure 1).
Table I. Characteristics of included patients.
Table II. Biomarker model based on LASSO regression when the reference model included age.
LASSO: Least absolute shrinkage and selection operator. *Comparison of the reference model including age with the models with added biomarker(s).
Figure 1. Comparison of the reference model and models with added biomarkers. A three-biomarker model including NT-3, GPNMB, and MSLN with age as the reference model showed the highest prediction of survival (AUC=0.792, p=0.004).
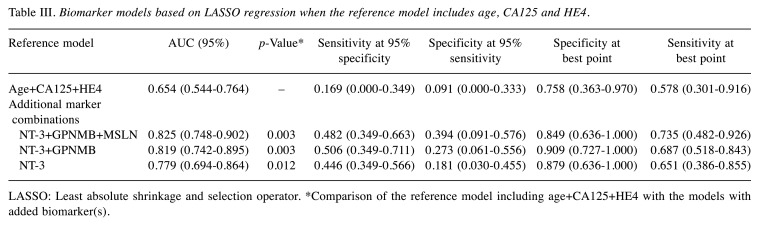
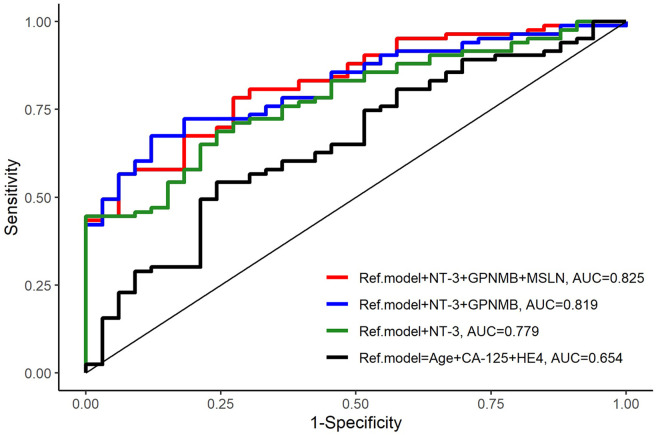
Adding CA125 and HE4 to age in the reference model improved the results [AUC=0.654 (0.544-0.764)], and the performance of CA125 and HE4 was further improved by the addition of NT-3+GPNMB+MSLN [AUC=0.825 (0.748-0.902); p= 0.003] (Table III, Figure 2).
Table III. Biomarker models based on LASSO regression when the reference model includes age, CA125 and HE4.
LASSO: Least absolute shrinkage and selection operator. *Comparison of the reference model including age+CA125+HE4 with the models with added biomarker(s).
Figure 2. Comparison of the reference model and models with added biomarkers. A three-biomarker model including NT-3+GPNMB+MSLN with age+CA-125+HE4 as the reference model showed the highest prediction of survival (AUC=0.825, p=0.003).
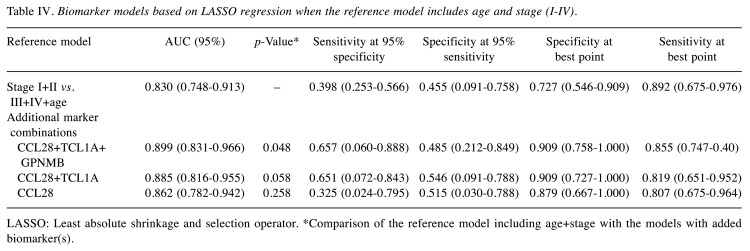
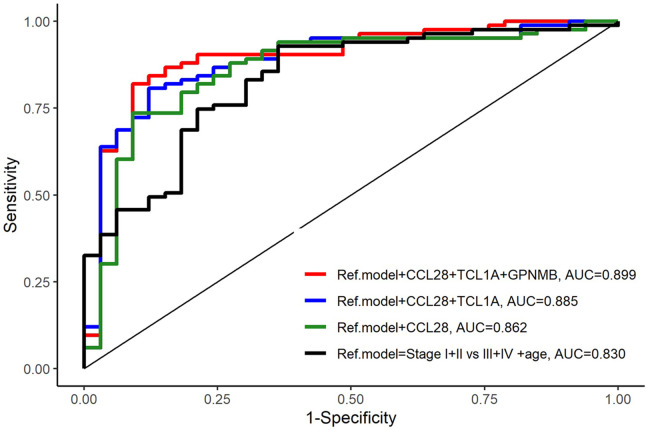
A third reference model was tested including age and stage [early (stage I+II) or late (stage III+IV), AUC=0.830 (0.748-0.913)]. The addition of the CCL28+TCL1A+GPNMB biomarkers to this reference model was found to create the best model for predicting OS [AUC=0.899 (0.831-0.966); p=0.048] (Table IV, Figure 3). When one or both biomarkers (GPNMB and TCL1A) were removed from this model, no statistical significance was observed (p=0.058 and p=0.258).
Table IV. Biomarker models based on LASSO regression when the reference model includes age and stage (I-IV).
LASSO: Least absolute shrinkage and selection operator. *Comparison of the reference model including age+stage with the models with added biomarker(s).
Figure 3. Comparison of the reference model and models with added biomarkers. A three-biomarker model including CCL28+TCL1A+GPNMB with Stage I+II vs. III+IV+age as the reference model showed the highest prediction of survival (AUC=0.899, p=0.048).
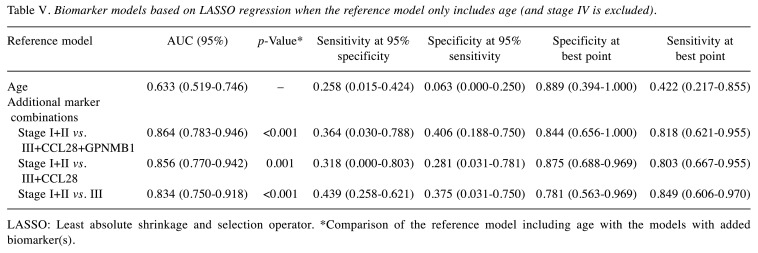
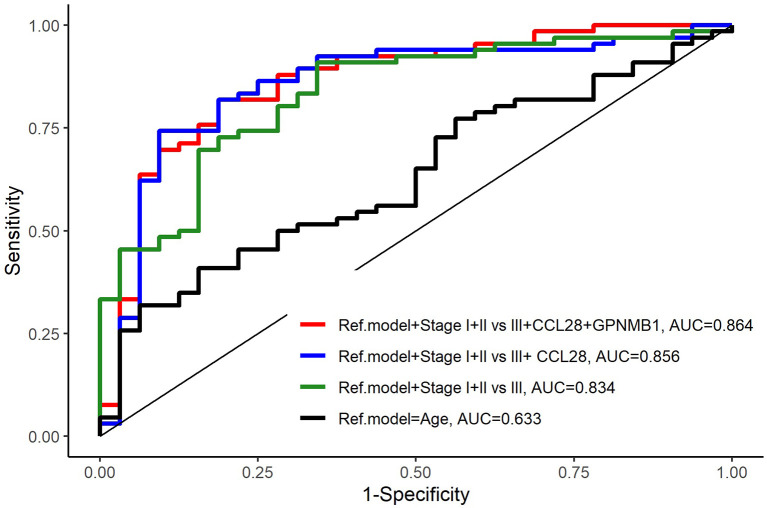
Finally, starting from a reference model using age only, the addition of stage (early stage I+II vs. late stage III) and CCL28+GPNMB biomarkers improved the prediction of OS and was statistically significant in all combinations, with the best performance for the model including age, stage and CCL28+GPNMB [AUC=0.864 (0.783-0.946); p<0.00] (Table V, Figure 4).
Table V. Biomarker models based on LASSO regression when the reference model only includes age (and stage IV is excluded).
LASSO: Least absolute shrinkage and selection operator. *Comparison of the reference model including age with the models with added biomarker(s).
Figure 4. Comparison of the reference model and models with added biomarkers. The model including stage I+II vs. III+CCL28+GPNMB1 with age as the reference model showed the highest prediction of survival (AUC=0.864, p<0.001).
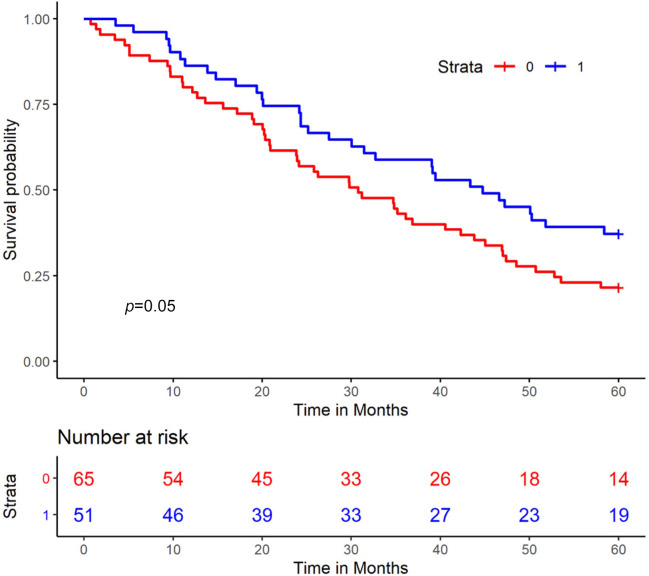
The combination of high levels of both CA125 and HE4 (both biomarkers divided at median) predicted poorer survival (p=0.05) (Figure 5).
Figure 5. Kaplan-Meier analysis of overall survival in patients in terms of CA125 and HE4 levels. Patients with both values above the median are shown in red, and patients with one or both biomarkers below the median are shown in blue.
Discussion
In this explorative study evaluating the performance of plasma protein biomarkers for predicting OS in EOC patients, we found that biomarker panels can improve the prediction of survival. The model including age and the combination of NT-3, GPNMB, MSLN, CA125 and HE4 was the best model to predict overall survival.
Recently, neurotrophins such as NT-3 have been shown to regulate angiogenesis through direct and indirect mechanisms (Garrido et al. 2019) (13). Neurotrophins include nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), and neurotrophins 3 and 4/5 (NT-3, NT 4/5), which have a high affinity for tropomyosin kinase receptors (TRKs) that regulate the development and plasticity of the nervous system and nonneuronal tissues, including reproductive organs such as the ovaries. Studies have shown that several types of cancer overexpress neurotrophins, which contribute to tumour progression and angiogenesis (13). The FDA and EMA have approved the use of pharmacologic inhibitors of pan-TRK receptors in patients with TRK fusion-positive cancers; however, thus far, they have not been used in ovarian cancer treatment.
The low sensitivity and limited specificity of CA125 to detect early-stage EOC (50-62%) do not fulfil the requirements to use CA125 as a screening biomarker in asymptomatic women (14). However, CA125 in combination with other biomarkers or supplemental data, such as age or disease stage in our study, or ultrasound markers can improve performance. The ROMA and ADNEX algorithms, especially in premenopausal women (14), can help differentiate benign ovarian tumours from cancer (15,16). According to our results, the combination of high levels of CA125 and HE4 with the addition of NT-3 showed worse five-year OS.
The risk of EOC as well as other cancers increases with age (17,18). Combining age with the already well-known markers CA125 and HE4 and the addition of NT-3 improved the prediction model of OS significantly. By adding GPNMB and MSLN, we expanded the study model, but no statistically significant results were found, which may have been influenced by the low numbers of patients in the study sample.
We found that increased levels of glycoprotein non-metastatic B (GPNMB) predicted unfavourable survival. GPNMB is also known as osteoactivin, a transmembrane protein overexpressed in several cancer tissues, such as breast cancer (19,20), stomach cancer (21), colorectal cancer (22), hepatocellular cancer (23) and others. Higher GPNMB levels have been demonstrated to promote angiogenesis, migration, invasion, and metastasis of cancer cells (24,25).
In our models, mesothelin (MSLN) was also a biomarker of worse survival. The expression of MSLN is dysregulated in several types of cancer, including pancreatic cancer (26) and ovarian cancer (27). Studies have shown that abnormal expression of MSLN plays an important role in tumour cell growth, invasion, and metastasis (28,29). In ovarian cancer, the specific binding of MSLN and CA125 can mediate the adhesion of tumour cells, which promotes the implantation and metastasis of ovarian cancer in the pelvic and abdominal cavities (30).
High levels of CCL28 in combination with TCL1A and GPNMB as well as EOC stage and age implied worse survival in our study. CCL28 has an important role in regulating the chemotaxis of cells (31).
The results of this study should be interpreted with caution due to the heterogeneity of the epithelial subtypes of histopathology, as only patients scheduled for primary upfront surgery were included in the study, and no patients scheduled for neoadjuvant chemotherapy were included. Cross-validation was used to validate the results. Despite the limitations of the study sample, we obtained positive results, which can provide an impetus for further research.
Conclusion
In summary, we identified biomarker panels predicting overall survival in EOC patients. The model including age and the combination of neurotrophin-3 (NT-3)+trans-membrane glycoprotein NMB (GPNMB)+mesothelin (MSLN)+cancer antigen 125 (CA125) and human epididymis protein 4 (HE4) was the best model to predict overall survival. Future research is warranted to replicate our results and identify additional biomarkers in panels that can predict the prognosis of EOC patients.
Funding
The study was supported by funds from the Swedish Cancer Foundation and Regional Funds Region Skåne. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of Interest
The Authors declare that they have no conflicts of interest.
Authors’ Contributions
AD: Study conception and design; data acquisition, analysis, and interpretation; drafting and revising the manuscript. AÅ: Data acquisition and interpretation; drafting and revising the manuscript. PL: Data acquisition and revising the manuscript. CB: Study conception and design; data acquisition, analysis, and interpretation; revising the manuscript.
Acknowledgements
We are grateful to all the women who participated in this study; to the staff at the Department of Obstetrics and Gynecology, Lund Hospital, for assistance with collecting the blood samples; and to the laboratory staff at the Division of Oncology, Medicon Village, Lund University, for help with all practical issues concerning the preparation and shipping of samples.
References
- 1.Kalapotharakos G, Högberg T, Bergfeldt K, Borgfeldt C. Long-term survival in women with borderline ovarian tumors: a population-based survey of borderline ovarian tumors in Sweden 1960-2007. Acta Obstet Gynecol Scand. 2016;95(4):473–479. doi: 10.1111/aogs.12846. [DOI] [PubMed] [Google Scholar]
- 2.Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2017;41:3–14. doi: 10.1016/j.bpobgyn.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Leandersson P, Kalapotharakos G, Henic E, Borgfeldt H, Petzold M, Høyer-Hansen G, Borgfeldt C. A biomarker panel increases the diagnostic performance for epithelial ovarian cancer type I and II in young women. Anticancer Res. 2016;36(3):957–965. [PubMed] [Google Scholar]
- 4.Paek J, Lee SH, Yim GW, Lee M, Kim YJ, Nam EJ, Kim SW, Kim YT. Prognostic significance of human epididymis protein 4 in epithelial ovarian cancer. Eur J Obstet Gynecol Reprod Biol. 2011;158(2):338–342. doi: 10.1016/j.ejogrb.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 5.Kalapotharakos G, Asciutto C, Henic E, Casslén B, Borgfeldt C. High preoperative blood levels of HE4 predicts poor prognosis in patients with ovarian cancer. J Ovarian Res. 2012;5(1):20. doi: 10.1186/1757-2215-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batchu RB, Gruzdyn OV, Kolli BK, Dachepalli R, Umar PS, Rai SK, Singh N, Tavva PS, Weaver DW, Gruber SA. IL-10 signaling in the tumor microenvironment of ovarian cancer. Adv Exp Med Biol. 2021;1290:51–65. doi: 10.1007/978-3-030-55617-4_3. [DOI] [PubMed] [Google Scholar]
- 7.Lane D, Matte I, Garde-Granger P, Bessette P, Piché A. Ascites IL-10 promotes ovarian cancer cell migration. Cancer Microenviron. 2018;11(2-3):115–124. doi: 10.1007/s12307-018-0215-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matte I, Lane D, Laplante C, Rancourt C, Piché A. Profiling of cytokines in human epithelial ovarian cancer ascites. Am J Cancer Res. 2012;2(5):566–580. [PMC free article] [PubMed] [Google Scholar]
- 9.Han GH, Yun H, Chung JY, Kim JH, Cho H. TMED9 expression level as a biomarker of epithelial ovarian cancer progression and prognosis. Cancer Genomics Proteomics. 2022;19(6):692–702. doi: 10.21873/cgp.20352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leandersson P, Åkesson A, Hedenfalk I, Malander S, Borgfeldt C. A multiplex biomarker assay improves the diagnostic performance of HE4 and CA125 in ovarian tumor patients. PLoS One. 2020;15(10):e0240418. doi: 10.1371/journal.pone.0240418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Assarsson E, Lundberg M, Holmquist G, Björkesten J, Thorsen SB, Ekman D, Eriksson A, Rennel Dickens E, Ohlsson S, Edfeldt G, Andersson AC, Lindstedt P, Stenvang J, Gullberg M, Fredriksson S. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9(4):e95192. doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The R Project: R project for statistical computing, 2022. Available at: https://www.r-project.org. [Last accessed on February 19, 2023]
- 13.Garrido MP, Torres I, Vega M, Romero C. Angiogenesis in gynecological cancers: role of neurotrophins. Front Oncol. 2019;9:913. doi: 10.3389/fonc.2019.00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sölétormos G, Duffy MJ, Othman Abu Hassan S, Verheijen RH, Tholander B, Bast RC Jr, Gaarenstroom KN, Sturgeon CM, Bonfrer JM, Petersen PH, Troonen H, CarloTorre G, Kanty Kulpa J, Tuxen MK, Molina R. Clinical use of cancer biomarkers in epithelial ovarian cancer: updated guidelines from the European Group on Tumor Markers. Int J Gynecol Cancer. 2016;26(1):43–51. doi: 10.1097/IGC.0000000000000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bandiera E, Romani C, Specchia C, Zanotti L, Galli C, Ruggeri G, Tognon G, Bignotti E, Tassi RA, Odicino F, Caimi L, Sartori E, Santin AD, Pecorelli S, Ravaggi A. Serum human epididymis protein 4 and risk for ovarian malignancy algorithm as new diagnostic and prognostic tools for epithelial ovarian cancer management. Cancer Epidemiol Biomarkers Prev. 2011;20(12):2496–2506. doi: 10.1158/1055-9965.EPI-11-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furrer D, Grégoire J, Turcotte S, Plante M, Bachvarov D, Trudel D, Têtu B, Douville P, Bairati I. Performance of preoperative plasma tumor markers HE4 and CA125 in predicting ovarian cancer mortality in women with epithelial ovarian cancer. PLoS One. 2019;14(6):e0218621. doi: 10.1371/journal.pone.0218621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rooth C. Ovarian cancer: risk factors, treatment and management. Br J Nurs. 2013;22(17):S23–S30. doi: 10.12968/bjon.2013.22.Sup17.S23. [DOI] [PubMed] [Google Scholar]
- 18.Doubeni CA, Doubeni AR, Myers AE. Diagnosis and management of ovarian cancer. Am Fam Physician. 2016;93(11):937–944. [PubMed] [Google Scholar]
- 19.Rose AA, Pepin F, Russo C, Abou Khalil JE, Hallett M, Siegel PM. Osteoactivin promotes breast cancer metastasis to bone. Mol Cancer Res. 2007;5(10):1001–1014. doi: 10.1158/1541-7786.MCR-07-0119. [DOI] [PubMed] [Google Scholar]
- 20.Huang YH, Chu PY, Chen JL, Huang CT, Huang CC, Tsai YF, Wang YL, Lien PJ, Tseng LM, Liu CY. Expression pattern and prognostic impact of glycoprotein non-metastatic B (GPNMB) in triple-negative breast cancer. Sci Rep. 2021;11(1):12171. doi: 10.1038/s41598-021-91588-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rho HW, Lee BC, Choi ES, Choi IJ, Lee YS, Goh SH. Identification of valid reference genes for gene expression studies of human stomach cancer by reverse transcription-qPCR. BMC Cancer. 2010;10:240. doi: 10.1186/1471-2407-10-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eldai H, Periyasamy S, Al Qarni S, Al Rodayyan M, Muhammed Mustafa S, Deeb A, Al Sheikh E, Afzal M, Johani M, Yousef Z, Aziz MA. Novel genes associated with colorectal cancer are revealed by high resolution cytogenetic analysis in a patient specific manner. PLoS One. 2013;8(10):e76251. doi: 10.1371/journal.pone.0076251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian F, Liu C, Wu Q, Qu K, Wang R, Wei J, Meng F, Liu S, Chang H. Upregulation of glycoprotein nonmetastatic B by colony-stimulating factor-1 and epithelial cell adhesion molecule in hepatocellular carcinoma cells. Oncol Res. 2013;20(8):341–350. doi: 10.3727/096504013X13657689382851. [DOI] [PubMed] [Google Scholar]
- 24.Rose AA, Annis MG, Dong Z, Pepin F, Hallett M, Park M, Siegel PM. ADAM10 releases a soluble form of the GPNMB/Osteoactivin extracellular domain with angiogenic properties. PLoS One. 2010;5(8):e12093. doi: 10.1371/journal.pone.0012093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rich JN, Shi Q, Hjelmeland M, Cummings TJ, Kuan CT, Bigner DD, Counter CM, Wang XF. Bone-related genes expressed in advanced malignancies induce invasion and metastasis in a genetically defined human cancer model. J Biol Chem. 2003;278(18):15951–15957. doi: 10.1074/jbc.M211498200. [DOI] [PubMed] [Google Scholar]
- 26.Le K, Wang J, Zhang T, Guo Y, Chang H, Wang S, Zhu B. Overexpression of mesothelin in pancreatic ductal adenocarcinoma (PDAC) Int J Med Sci. 2020;17(4):422–427. doi: 10.7150/ijms.39012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Tian W, Zhang H, Zhang Z, Zhao Q, Chang L, Lei N, Zhang W. MSLN correlates with immune infiltration and chemoresistance as a prognostic biomarker in ovarian cancer. Front Oncol. 2022;12:830570. doi: 10.3389/fonc.2022.830570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avula LR, Rudloff M, El-Behaedi S, Arons D, Albalawy R, Chen X, Zhang X, Alewine C. Mesothelin enhances tumor vascularity in newly forming pancreatic peritoneal metastases. Mol Cancer Res. 2020;18(2):229–239. doi: 10.1158/1541-7786.MCR-19-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inoue S, Tsunoda T, Riku M, Ito H, Inoko A, Murakami H, Ebi M, Ogasawara N, Pastan I, Kasugai K, Kasai K, Ikeda H, Inaguma S. Diffuse mesothelin expression leads to worse prognosis through enhanced cellular proliferation in colorectal cancer. Oncol Lett. 2020;19(3):1741–1750. doi: 10.3892/ol.2020.11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coelho R, Marcos-Silva L, Ricardo S, Ponte F, Costa A, Lopes JM, David L. Peritoneal dissemination of ovarian cancer: role of MUC16-mesothelin interaction and implications for treatment. Expert Rev Anticancer Ther. 2018;18(2):177–186. doi: 10.1080/14737140.2018.1418326. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez MW, Paquet AC, Yang YH, Erle DJ. Differential gene expression by integrin beta 7+ and beta 7- memory T helper cells. BMC Immunol. 2004;5:13. doi: 10.1186/1471-2172-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]