Abstract
Observational data can be used to conduct drug surveillance and effectiveness studies, investigate treatment pathways, and predict patient outcomes. Such studies require developing executable algorithms to find patients of interest or phenotype algorithms. Creating reliable and comprehensive phenotype algorithms in data networks is especially hard as differences in patient representation and data heterogeneity must be considered. In this paper, we discuss a process for creating a comprehensive concept set and a recommender system we built to facilitate it. PHenotype Observed Entity Baseline Endorsements (PHOEBE) uses the data on code utilization across 22 electronic health record and claims datasets mapped to the Observational Health Data Sciences and Informatics (OHDSI) Common Data Model from the 6 countries to recommend semantically and lexically similar codes. Coupled with Cohort Diagnostics, it is now used in major network OHDSI studies. When used to create patient cohorts, PHOEBE identifies more patients and captures them earlier in the course of the disease.
Introduction
Studies using observational data such as electronic health records (EHR) and administrative claims have been shown to impact decision-making and improve patient outcomes [1]. While addressing some of the concerns raised regarding randomized clinical trials such as limited patient involvement, short follow-up and high cost, they pose additional challenges related to the use of observational data. The latter include underlying bias, missing and incomplete data and more [2]. One of the main challenges in observational research is the accurate capture of patients of interest and their history and diseases. To achieve that, researchers develop computable phenotyping algorithms that aim at transforming the raw EHR and claims data into clinically relevant features [3].
Our ability to create accurate and reliable phenotypes depends on two components: the ability to derive relevant and comprehensive sets of codes (or building blocks) that are used to code the corresponding patients in the data and the accuracy of the logic applied to those codes. While machine learning approaches have been leveraged to use the data in place of human logic (which can be biased), the issue of selecting appropriate codes remains largely unsolved in the setting of moving learned phenotypes among disparate institutions without retraining.
Common approaches for code selection include deriving codes from published phenotypes or using local expert knowledge [4,5]. Nevertheless, as has been shown, phenotype definitions and code sets are not readily transportable to other institutions and are characterized by lower performance when applied to other data sources [6].
Another common approach is to use machine learning approaches like multi-view banded spectral clustering [7], non-negative tensor-factorization [8], latent Dirichlet allocation [9], embeddings with knowledge graphs and EHR [10] to derive the codes from the structured data or supplementing the latter with the extracts from unstructured data [11,12]. While it potentially eliminates human subjectivity, learned knowledge is still limited to the institution the model was developed in and therefore may not be applicable to other institutions [13].
This challenge becomes especially hard when a study is conducted in a distributed network. Large-scale observational studies in distributed networks leverage large patient populations to produce reliable evidence, but they require recognition and reconciliation of differences in patient data capture. As we observed previously, data sources are highly heterogeneous with a large proportion of the codes being uniquely used in one institution but not in the others [14]. Researchers also need to account for variation in the granularity of codes. For example, non-US data sources code events using more broad general codes compared to the US data sources [15].
Recent Observational Health Data Sciences and Informatics (OHDSI) Observational Medical Outcomes Partnership (OMOP) studies [4] enable leveraging a shared knowledge base of concepts and their utilization across the network to construct comprehensive concept sets and phenotypes. In this work, we will describe a recommender system for code selection created based on this knowledge base.
Methods
Concept Prevalence Study
In this study, we used the data set generated in the Concept Prevalence study [15]. There, we collected clinical codes (procedures, conditions, laboratory tests, vital signs and other observations) along with the frequency of their use from 22 data sources across six countries (US, Japan, Korea, Australia, Germany, France). All data sources were transformed into the OMOP Common Data Model version 5 [16] and both the codes used in the source and the codes standardized to the OMOP Standardized Vocabularies were collected. The frequency of codes less than 100 was rounded to 100. Here, we will use the term ‘clinical code’ and ‘concept’ interchangeably as a representation of a term and its code in an ontology.
PHOEBE development
The initial set used by our recommender system - PHenotype Observed Entity Baseline Endorsements (PHOEBE) - consisted of eleven million unique concepts appearing in at least one source within the network, with 272 billion records summarized. Each concept therefore had an aggregated estimate of the frequency of its use across all the data sources as well as an aggregated estimate of the frequency of use of all its descendants. The latter was derived using the OMOP Standardized Vocabularies and represented the aggregated frequency of all child concepts in a corresponding hierarchy (RxNorm, SNOMED, LOINC etc.). For the concepts that do not participate in the OMOP Standardized Vocabularies hierarchy (such as ICD10-CM, Read or NDC), the estimate of descendants’ frequency was equal to the aggregated frequency of the code itself.
Aggregated frequencies were used to pre-compute a set of recommended terms for all standard concepts in the OMOP Standardized Vocabularies. First, string matching (for concepts and their synonyms) was used to select all lexically similar concepts for each standard concept. Then, semantically similar concepts were added by selecting the most proximal concepts within an ontology and through the crosswalks between the adjacent ontologies. These included both the concepts proximal and distal to the common ancestor as well as those belonging to a different hierarchy sub-tree (no common ancestor).
The concepts were subsequently filtered to those that provide an adequate data source capture. That was achieved by eliminating the concepts from those ontologies that cover less than half of the data sources (either directly or through mapping from the source ontologies) in a given domain. We also removed the concepts not used in any data source. Finally, we removed the broad terms that are not likely to contribute to a phenotype definition (such as ‘Disorder’ or ‘Family History of Disease’).
PHOEBE application
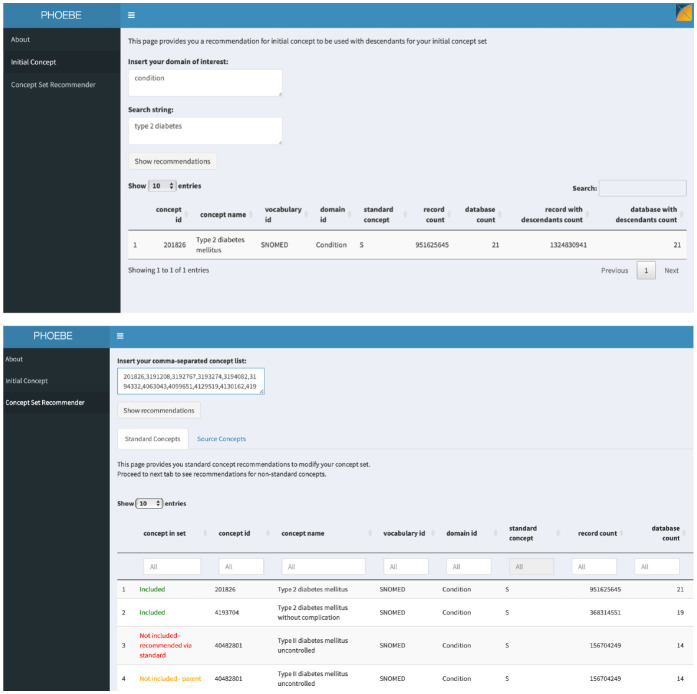
We created an R Shiny-based application available publicly (https://data.ohdsi.org/PHOEBE). It has two parts: initial concept recommender and concept set recommender (Figure 1).
Figure 1.
PHOEBE R Shiny application: initial concept and concept set recommender.
For the initial concept, a user inputs a string that specifies her clinical idea (such as diabetes mellitus type 2) and PHOEBE outputs the best match for a code that represents this clinical idea prioritized based on the number of data sources covered and an aggregated frequency of both the code itself and its descendants. The user can then use this code with all its descendants as a starting place for developing the concept set.
Concept set recommender takes a string of codes (obtained in the previous step) and outputs a set of codes divided into the following groups:
- included codes
- not included concepts (parent and child codes of the codes included in the concept set)
- recommended codes (recommended through matching in source vocabularies or standard vocabularies).
The output is again prioritized based on the aggregated frequency to focus decision-making on the concepts that provide the largest gain in record count.
Evaluation
For validation, we used an electronic health record data source (Columbia University Irving Medical Center EHR) and two administrative claims data sources (IBM MarketScan® Multi-State Medicaid Database and IBM
MarketScan® Medicare Supplemental Database) translated to OMOP Common Data Model. We selected diabetes mellitus type II, diabetes mellitus type I, heart failure and attention deficit hyperactivity disorder (ADHD) – common conditions that were extensively studied in the observational literature and for which drug treatment exists. For each of the conditions, PHOEBE was used to create concept sets following the steps outlined above.
As a reference, we selected two eMERGE network phenotypes representing the same clinical ideas [17]. eMERGE is a national network for high-throughput genetic research that developed and deployed numerous electronic phenotype algorithms. Phenotypes have been created by highly qualified multidisciplinary teams and often been taking up to 6–8 months to develop and validate. For each eMERGE phenotype, we extracted ICD9-CM concept sets used in the original implementation and translated them to SNOMED-CT concepts [6]. We then created patient cohorts by selecting patients with at least one occurrence of a diagnosis code from corresponding concept sets and with at least 365 days of prior observation to ensure data coverage. For each cohort, we followed patients to look for a specific treatment, which included any occurrence of insulin products for type I diabetes mellitus; oral antidiabetic drugs (metformin, sulfonylureas, thiazolidinediones, dipeptidyl peptidase IV inhibitors and glucagon-like peptide-1 agonists) for type II diabetes mellitus; beta blockers, angiotensin-converting enzyme inhibitors and diuretics for heart failure and viloxazine, atomoxetine, amphetamine methylphenidate and guanfacine for ADHD [19–22].
We computed the positive predictive value (PPV) of each phenotype, which was defined as a proportion of people with a diagnostic code who also had subsequent treatment with corresponding drugs. Additionally, for patients with subsequent treatment identified by both PHOEBE and eMERGE concept set-based algorithms, condition index dates (the date a disease was first observed in a patient) were extracted and compared.
Results
Over the past year, PHOEBE has been used in all major network studies conducted in the OHDSI Network. They include clinical studies such as characterizing patients with COVID-19 [23–28] and methodological studies such as investigating the sensitivity of background rates or validation of prediction models [29,30]. PHOEBE is continuously used for new studies such as Large-scale Evidence Generation and Evaluation across a Network of Databases (LEGEND) for type 2 diabetes mellitus [31]. The studies were published in high-impact journals like BMJ, Nature Communications, Lancet Rheumatology, Rheumatology and others.
As the abovementioned studies involved analyzing the data from up to 25 US and international data sources, PHOEBE was required to generate comprehensive concept sets that would reflect the codes used in all of the data sources.
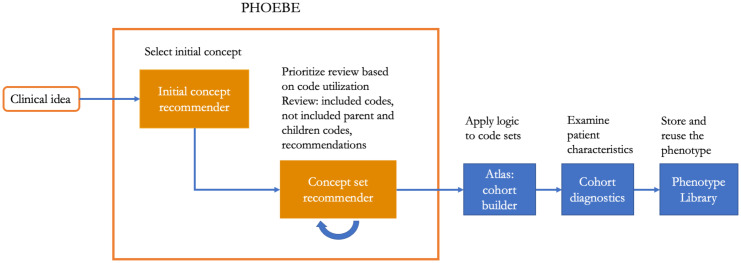
Coupled with the tools for examining patient cohorts (Cohort Diagnostics, Figure 2), it provided an opportunity to systematically examine and refine phenotype definitions used for defining target, comparator groups or outcomes of interest.
Figure 2.
OHDSI pipeline for developing, evaluating and storing phenotypes, including PHOEBE, ATLAS, Cohort Diagnostics and Phenotype Library.
Evaluation
While the main benefit of PHOEBE is providing the data to support researchers’ decision making, we also formally evaluated its impact on patient selection.
When looking at the cohorts created using the concept sets provided by PHOEBE and used in the eMERGE phenotypes, we found that on average PHOEBE identified more patients while preserving similar positive predictive value (Table 1). We observed high heterogeneity in the magnitude of patient gain among data sources and conditions. For example, for type I diabetes, the algorithm created with PHOEBE identified approximately the same set of patients as the algorithm that used well-curated concept set from eMERGE. Notably, the cohort of type II diabetes patients created using PHOEBE had more than 5 times more patients and had a higher positive predictive value.
Table 1.
Comparison of eMERGE and PHOEBE concept set-based algorithms’ performance. PPV – positive predictive value, CUIMC – Columbia University Irving Medical Center EHR, MDCR - IBM MarketScan® Medicare Supplemental Database, MDCD - IBM MarketScan® Multi-State Medicaid Database.
| eMERGE cohort | PHOEBE cohort | Cohort overlap | ||||||
|---|---|---|---|---|---|---|---|---|
| Patients with subsequent treatment | Total | PPV | Patients with subsequent treatment | Total | PPV | |||
| Diabetes mellitus type I | CUIMC | 7,599 | 25,701 | 0.34 | 7,659 | 25,884 | 0.34 | 25,635 |
| MDCD | 25,954 | 191,710 | 0.13 | 26,240 | 194,100 | 0.13 | 189,810 | |
| MDCR | 19,274 | 185,874 | 0.10 | 18,684 | 181,712 | 0.11 | 179,236 | |
| Diabetes mellitus type II | CUIMC | 42,200 | 211,904 | 0.20 | 44,828 | 218,127 | 0.21 | 211,903 |
| MDCD | 342,975 | 1,807,688 | 0.19 | 383,435 | 1,849,177 | 0.21 | 1,807,019 | |
| MDCR | 105,449 | 386,265 | 0.27 | 804,415 | 2,172,925 | 0.37 | 386,237 | |
| ADHD | CUIMC | 5,922 | 29,890 | 0.20 | 7,002 | 35,291 | 0.20 | 29,890 |
| MDCD | 722,485 | 1,471,559 | 0.49 | 809,093 | 1,657,347 | 0.49 | 1,471,542 | |
| MDCR | 7,158 | 17,065 | 0.42 | 7,875 | 18,535 | 0.43 | 17,065 | |
| Heart Failure | CUIMC | 71,281 | 162,297 | 0.44 | 74,121 | 168,111 | 0.44 | 162,281 |
| MDCD | 332,929 | 870,080 | 0.38 | 342,112 | 893,807 | 0.38 | 869,500 | |
| MDCR | 892,689 | 1,166,980 | 0.76 | 919,077 | 1,204,488 | 0.76 | 1,165,900 | |
In general, patient gain was less notable in CUIMC, which can be explained by the fact that eMERGE phenotypes were partially developed on CUIMC data. Even when there was no significant difference in PPV, PHOEBE’s algorithm identified more patients with subsequent treatment.
We obtained similar performance upon repeating the procedure for other randomly selected versions of these phenotypes found in the literature.
Aside from evaluating the ability of PHOEBE to identify patients of interest, we also examined differences in the index dates (date of first observation of a disease in a data source) for the patients identified by both PHOEBE and eMERGE concept set-based algorithms. We observed that PHOEBE can identify patients earlier on in the course of the disease (Table 2).
Table 2.
Comparison of condition onset date (index date) in patients with different index dates identified by both eMERGE and PHOEBE concept set-based algorithms. CUIMC – Columbia University Irving Medical Center EHR, MDCR - IBM MarketScan® Medicare Supplemental Database, MDCD - IBM MarketScan® Multi-State Medicaid Database.
| MDCD | MCDR | CUMC | ||||
| Patients, n | Difference in days, median (IQR) | Patients, n | Difference in days, median (IQR) | Patients, n | Difference in days, median (IQR) | |
| Diabetes mellitus type I | 2,243 | 371 (139-795) | 2,807 | 407 (176 - 894) | 206 | 1,001 (206-1,915) |
| Diabetes mellitus type II | 253,728 | 77 (22-226) | 957 | 122 (13-417) | 11,536 | 189 (28-873) |
| ADHD | 165,590 | 201 (59-525) | 203 | 175 (61-318) | 1,801 | 146 (35-532) |
| Heart Failure | 16,941 | 49 (3-298) | 25,294 | 107 (6-472) | 2,242 | 78 (6-724) |
For example, patients with different onset dates, on average, presented with diabetes mellitus type I more than two years earlier when using the PHOEBE algorithm in CUMC and more than one year earlier in MDCD and MDCR. This can be explained by multiple patients having unspecified diabetes mellitus codes prior to being treated by endocrinologists. This pattern was consistently observed in all conditions and data sources, which may reflect that the use of broad non-specific codes by clinicians is common in early stages of disease. This finding suggests that the use of concept sets consisting of only narrow specific codes may introduce index even misspecification as such codes may capture patients not at the first observation of the disease but later on in the course.
Examples of the codes recommended through PHOEBE included SNOMED-CT codes “Hyperosmolar coma due to type 1 diabetes mellitus”, “Type 1 diabetes mellitus uncontrolled “, “Peripheral circulatory disorder due to type 1 diabetes mellitus”, “Disorder of nervous system due to type 1 diabetes mellitus” and other complications for diabetes mellitus type I. For diabetes mellitus type II, PHOEBE also recommended complications of diabetes along with broad terms such as “Diabetic - poor control”. While the latter concept is broad, it accounts for 833,654 records in the Network. Given higher prevalence of type II diabetes compared to type I and high utilization of this code, researchers may want to use this code in their phenotype with additional restrictions like older age, absence of codes for type I diabetes or others.
Discussion
Selecting of relevant codes for a phenotype is not a trivial task, especially when conducting a study on multiple data sources. One of the most common approaches is borrowing concept sets from the previously published studies. Yet, as widely acknowledged, the performance on one data source does not guarantee similar performance on another.
For single-center studies, the researchers can get away with leveraging local data or expert knowledge to produce concept sets. The former approach was also advertised for organizing data models that do not involve semantic standardization, such as Sentinel or PCORNet [4,32]. While expert knowledge is critical in both phenotype development and evaluation, clinicians are oftentimes not familiar with the data or code use patterns [33]. While they may tend to extrapolate their practices to the other clinicians, real-world patterns of treatment are highly heterogeneous and variable [34].
Moreover, for published observational research the choices made for concept sets are rarely described. The readers can assess neither how the concept sets were arrived at, nor the implications of the choices made. We have no reason to believe that different institutions or individuals have the same approach to concept set selection, which means that a concept set, phenotype and, in turn, study results can be different when another researcher performs a selection for a given study. Using purely data-driven approaches with sufficient performance eliminates variability induced by an individual but provides little help when a study needs to be executed in a network. Moreover, the current large-scale initiatives that have a potential to impact decision-making are limited to specific domains or conditions and take years to develop and implement locally [35,36].
An intermediate between expert-based and data-driven approaches is a data-augmented expert-based approach, which uses the data to guide decision-making. There are several studies on describing how the data was used to learn the patterns for code use and guide clinicians and informaticians in concept set selection [37,38]. While providing important insights, they are limited to a single center or condition of interest and do not provide general guidance on how to create a concept set. Moreover, they do not describe the process for international networks that use disjoint ontologies to code their data.
This work benefits from the OHDSI Common Data Model that standardizes both the format and the content. The standards for computable phenotypes like Desiderata highlight the importance of the standardization of representation, use of common terminologies and data models for reproducible and reliable phenotypes [39,40]. In federated networks, their use is critical for timely execution, quality assurance and reproducibility.
Yet, it requires recognition and reconciliation of differences in patient representations across the network. PHOEBE leverages the code usage patterns to provide clinicians and informaticians with the data to ground their code selection decisions. While the final choice of including or excluding a code in the concept set is left to the researchers’ discretion, having the data allows them to both inspect the included codes and browse excluded yet relevant codes. Moreover, it allows prioritizing the inspection of the most used codes among hundreds of similar codes in the US and international ontologies.
Limitations
One of the limitations of this work is using only the US data sources for evaluation. As PHOEBE leverages data from a large network, it may show better performance on non-US data sources, which is yet to be shown. We used specific treatment as a proxy for patients being a true positive case. While imperfect, it allows comparing algorithm performance in absence of reliable methods to validate phenotypes in administrative claims data.
Conclusion
Patient phenotyping in network studies is time-consuming and oftentimes produces inconsistent results due to different patient representations in disparate data sources. We developed a recommender system that facilitates phenotype development standardization and comprehensive concept set creation. When PHOEBE is used, phenotyping algorithms identify more patients and capture them earlier in the course of disease.
Funding
This project was supported by the US National Library of Medicine grant R01 LM006910.
Figures & Table
References
- 1.Dreyer NA, Tunis SR, Berger M, et al. Why Observational Studies Should Be Among The Tools Used In Comparative Effectiveness Research. Health Affairs. 2010;29:1818–25. doi: 10.1377/hlthaff.2010.0666. doi:10.1377/hlthaff.2010.0666. [DOI] [PubMed] [Google Scholar]
- 2.Holve E, Lopez MH, Scott L, et al. A tall order on a tight timeframe: stakeholder perspectives on comparative effectiveness research using electronic clinical data. J Comp Eff Res. 2012;1:441–51. doi: 10.2217/cer.12.47. doi:10.2217/cer.12.47. [DOI] [PubMed] [Google Scholar]
- 3.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. Journal of the American Medical Informatics Association. 2013;20:117–21. doi: 10.1136/amiajnl-2012-001145. doi:10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc. 2016;23:1046–52. doi: 10.1093/jamia/ocv202. doi:10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. Journal of the American Medical Informatics Association. 2014;21:221–30. doi: 10.1136/amiajnl-2013-001935. doi:10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross MK, Zhu B, Natesan A, et al. B52. PEDIATRIC ASTHMA AND ALLERGY. American Thoracic Society; 2020. Accuracy of Asthma Computable Phenotypes to Identify Pediatric Asthma Cases at an Academic Institution; p. A3710–A3710. doi:10.1164/ajrccm-conference.2020.201.1_MeetingAbstracts.A3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, Zhang Y, Cai T, et al. Automated grouping of medical codes via multiview banded spectral clustering. Journal of Biomedical Informatics. 2019;100:103322. doi: 10.1016/j.jbi.2019.103322. doi:10.1016/j.jbi.2019.103322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Zhang Y, Schlueter DJ, et al. Detecting time-evolving phenotypic topics via tensor factorization on electronic health records: Cardiovascular disease case study. Journal of Biomedical Informatics. 2019;98:103270. doi: 10.1016/j.jbi.2019.103270. doi:10.1016/j.jbi.2019.103270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen P, Dong W, Lu X, et al. Deep representation learning for individualized treatment effect estimation using electronic health records. Journal of biomedical informatics. 2019;100:103303. doi: 10.1016/j.jbi.2019.103303. [DOI] [PubMed] [Google Scholar]
- 10.Lee J, Liu C, Kim JH, et al. Comparative effectiveness of medical concept embedding for feature engineering in phenotyping. JAMIA Open. 2021;4:ooab028. doi: 10.1093/jamiaopen/ooab028. doi:10.1093/jamiaopen/ooab028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu S, Chakrabortty A, Liao KP, et al. Surrogate-assisted feature extraction for high-throughput phenotyping. Journal of the American Medical Informatics Association. 2017;24:e143–9. doi: 10.1093/jamia/ocw135. doi:10.1093/jamia/ocw135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong C, Rush E, Liu M, et al. Clinical knowledge extraction via sparse embedding regression (KESER) with multi-center large scale electronic health record data. npj Digit Med. 2021;4:151. doi: 10.1038/s41746-021-00519-z. doi:10.1038/s41746-021-00519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y. Opportunities and Challenges in Data-Driven Healthcare Research. MEDICINE & PHARMACOLOGY. 2018. doi:10.20944/preprints201806.0137.v1.
- 14.Ostropolets A. Concept Heterogeneity in the OHDSI Network. 2019. https://www.ohdsi.org/2019-us-symposium-showcase-19/
- 15.Ostropolets A. Characterizing database granularity using SNOMED-CT hierarchy. AMIA 2020 Proceedings. 2020. [PMC free article] [PubMed]
- 16.Hripcsak G, Duke JD, Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud Health Technol Inform. 2015;216:574–8. [PMC free article] [PubMed] [Google Scholar]
- 17.and The eMERGE Network. Gottesman O, Kuivaniemi H, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genetics in Medicine. 2013;15:761–71. doi: 10.1038/gim.2013.72. doi:10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hripcsak G, Levine ME, Shang N, et al. Effect of vocabulary mapping for conditions on phenotype cohorts. Journal of the American Medical Informatics Association. 2018;25:1618–25. doi: 10.1093/jamia/ocy124. doi:10.1093/jamia/ocy124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kagawa R, Kawazoe Y, Ida Y, et al. Development of Type 2 Diabetes Mellitus Phenotyping Framework Using Expert Knowledge and Machine Learning Approach. J Diabetes Sci Technol. 2017;11:791–9. doi: 10.1177/1932296816681584. doi:10.1177/1932296816681584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spratt SE, Pereira K, Granger BB, et al. Assessing electronic health record phenotypes against gold-standard diagnostic criteria for diabetes mellitus. Journal of the American Medical Informatics Association. 2017;24:e121–8. doi: 10.1093/jamia/ocw123. doi:10.1093/jamia/ocw123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blecker S, Katz SD, Horwitz LI, et al. Comparison of Approaches for Heart Failure Case Identification From Electronic Health Record Data. JAMA Cardiol. 2016;1:1014. doi: 10.1001/jamacardio.2016.3236. doi:10.1001/jamacardio.2016.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slaby I, Hain HS, Abrams D, et al. An electronic health record (EHR) phenotype algorithm to identify patients with attention deficit hyperactivity disorders (ADHD) and psychiatric comorbidities. J Neurodevelop Disord. 2022;14:37. doi: 10.1186/s11689-022-09447-9. doi:10.1186/s11689-022-09447-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prieto-Alhambra D, Kostka K, Duarte-Salles T, et al. Unraveling COVID-19: a large-scale characterization of 4.5 million COVID-19 cases using CHARYBDIS. 2021. [DOI] [PMC free article] [PubMed]
- 24.Burn E, You SC, Sena AG, et al. Deep phenotyping of 34,128 adult patients hospitalised with COVID-19 in an international network study. Nature communications. 2020;11:1–11. doi: 10.1038/s41467-020-18849-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recalde M, Roel E, Pistillo A, et al. Characteristics and outcomes of 627 044 COVID-19 patients with and without obesity in the United States, Spain, and the United Kingdom. Published Online First: 3 September 2020. doi:10.1101/2020.09.02.20185173. [DOI] [PMC free article] [PubMed]
- 26.Reyes C, Pistillo A, Fernández-Bertolín S, et al. Characteristics and outcomes of patients with COVID-19 with and without prevalent hypertension: a multinational cohort study. BMJ Open. 2021;11:e057632. doi: 10.1136/bmjopen-2021-057632. doi:10.1136/bmjopen-2021-057632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan EH, Sena AG, Prats-Uribe A, et al. COVID-19 in patients with autoimmune diseases: characteristics and outcomes in a multinational network of cohorts across three countries. Rheumatology. 2021;60:SI37–50. doi: 10.1093/rheumatology/keab250. doi:10.1093/rheumatology/keab250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morales DR, Ostropolets A, Lai L, et al. Characteristics and outcomes of COVID-19 patients with and without asthma from the United States, South Korea, and Europe. Journal of Asthma. 2022. pp. 1–11. doi:10.1080/02770903.2021.2025392. [DOI] [PubMed]
- 29.Reps J, Kim C, Williams R, et al. Can we trust the prediction model? Demonstrating the importance of external validation by investigating the COVID-19 Vulnerability (C-19) Index across an international network of observational healthcare datasets. 2020.
- 30.Ostropolets A, Li X, Makadia R, et al. Empirical evaluation of the sensitivity of background incidence rate characterization for adverse events across an international observational data network. Health Informatics. 2021. doi:10.1101/2021.06.27.21258701.
- 31.Khera R, Scheumie MJ, Lu Y, et al. RESEARCH PROTOCOL: Large-scale evidence generation and evaluation across a network of databases for type 2 diabetes mellitus. Endocrinology (including Diabetes Mellitus and Metabolic Disease) 2021. doi:10.1101/2021.09.27.21264139.
- 32.Schneeweiss S, Brown JS, Bate A, et al. Choosing Among Common Data Models for Real‐World Data Analyses Fit for Making Decisions About the Effectiveness of Medical Products. Clin Pharmacol Ther. 2020;107:827–33. doi: 10.1002/cpt.1577. doi:10.1002/cpt.1577. [DOI] [PubMed] [Google Scholar]
- 33.Lanes S, Brown JS, Haynes K, et al. Identifying health outcomes in healthcare databases: Identifying Health Outcomes. Pharmacoepidemiol Drug Saf. 2015;24:1009–16. doi: 10.1002/pds.3856. doi:10.1002/pds.3856. [DOI] [PubMed] [Google Scholar]
- 34.Hripcsak G, Ryan PB, Duke JD, et al. Characterizing treatment pathways at scale using the OHDSI network. Proceedings of the National Academy of Sciences. 2016;113:7329–36. doi: 10.1073/pnas.1510502113. doi:10.1073/pnas.1510502113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown JS, Maro JC, Nguyen M, et al. Using and improving distributed data networks to generate actionable evidence: the case of real-world outcomes in the Food and Drug Administration’s Sentinel system. Journal of the American Medical Informatics Association. 2020;27:793–7. doi: 10.1093/jamia/ocaa028. doi:10.1093/jamia/ocaa028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ball R, Robb M, Anderson S, et al. The FDA’s sentinel initiative-A comprehensive approach to medical product surveillance. Clin Pharmacol Ther. 2016;99:265–8. doi: 10.1002/cpt.320. doi:10.1002/cpt.320. [DOI] [PubMed] [Google Scholar]
- 37.Morley KI, Wallace J, Denaxas SC, et al. Defining Disease Phenotypes Using National Linked Electronic Health Records: A Case Study of Atrial Fibrillation. PLoS ONE. 2014;9:e110900. doi: 10.1371/journal.pone.0110900. doi:10.1371/journal.pone.0110900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmad FS, Ricket IM, Hammill BG, et al. Computable Phenotype Implementation for a National, Multicenter Pragmatic Clinical Trial: Lessons Learned From ADAPTABLE. Circ: Cardiovascular Quality and Outcomes. 2020;13 doi: 10.1161/CIRCOUTCOMES.119.006292. doi:10.1161/CIRCOUTCOMES.119.006292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mo H, Thompson WK, Rasmussen LV, et al. Desiderata for computable representations of electronic health records-driven phenotype algorithms. Journal of the American Medical Informatics Association. 2015;22:1220–30. doi: 10.1093/jamia/ocv112. doi:10.1093/jamia/ocv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chapman M, Mumtaz S, Rasmussen LV, et al. Desiderata for the development of next-generation electronic health record phenotype libraries. GigaScience. 2021;10:giab059. doi: 10.1093/gigascience/giab059. doi:10.1093/gigascience/giab059. [DOI] [PMC free article] [PubMed] [Google Scholar]