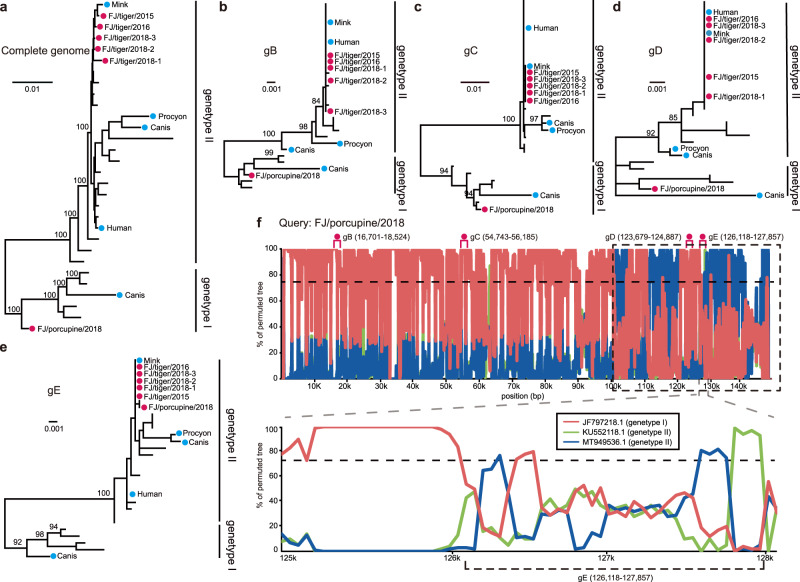
Fig. 5. Phylogenetic history and recombination in pseudorabies viruses (PRVs).
The phylogenetic trees were estimated based on a full genome sequences, and the b gB, (c) gC, d gD, and e gE genes, utilizing the best-fit nucleotide substitution model obtained by IQ-TREE (Minh et al., 2020). All trees were midpoint rooted. Numbers (>80) above branches are percentage bootstrap values for the major nodes. The scale bar represents the number of substitutions per site. Red circles indicate the six tiger- and porcupine-isolated PRVs generated in this study. f Similarity plot of the full-length PRV genome of porcupine PRV against sequences of a genotype I strain (JF797218.1) and two genotype II strains (KU552118.1 and MT949536.1). The parameters for the similarity plots are: window, 200 bp; step, 50 bp.