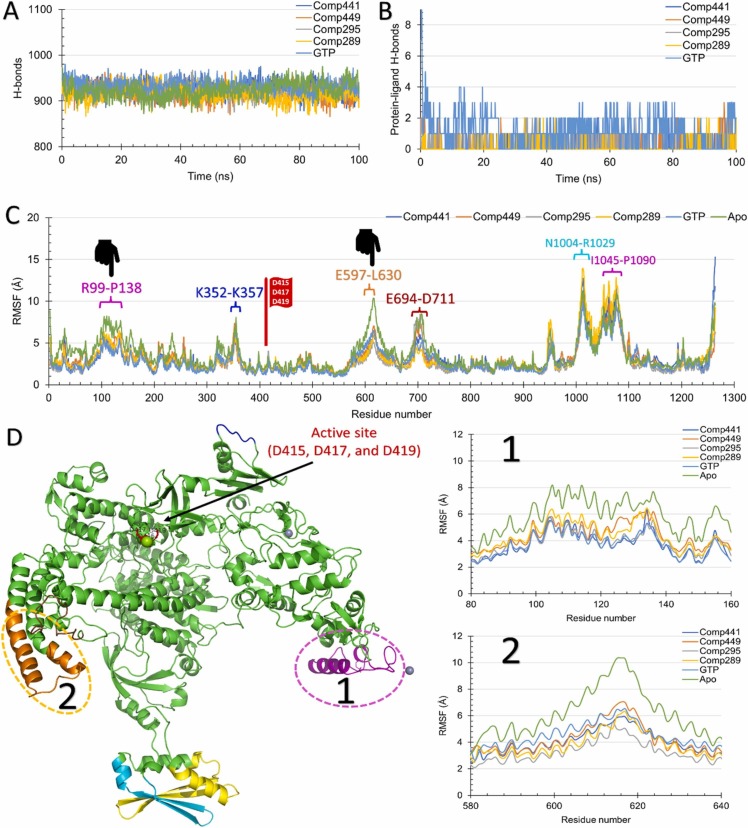
Fig. 5.
The molecular dynamics simulation analysis of the top four hits against hMPXV DdRp. (A) the total number of formed H-bonds, (B) protein-ligands formed H-bonds, versus the simulation time in ns for the four top hit complexes, the positive control (GTP), and the Apo form. (C) the per-residue root-mean-square fluctuations in Å are also shown for the same complexes. Highly fluctuating regions are indicated by different colors, while regions of reduced fluctuation compared to the apo DdRp are marked on the RMSF curve. (D) the two regions of reduced RMSF in the complexes compared to the apo DdRp are enlarged (right) and shown in the structure (left).