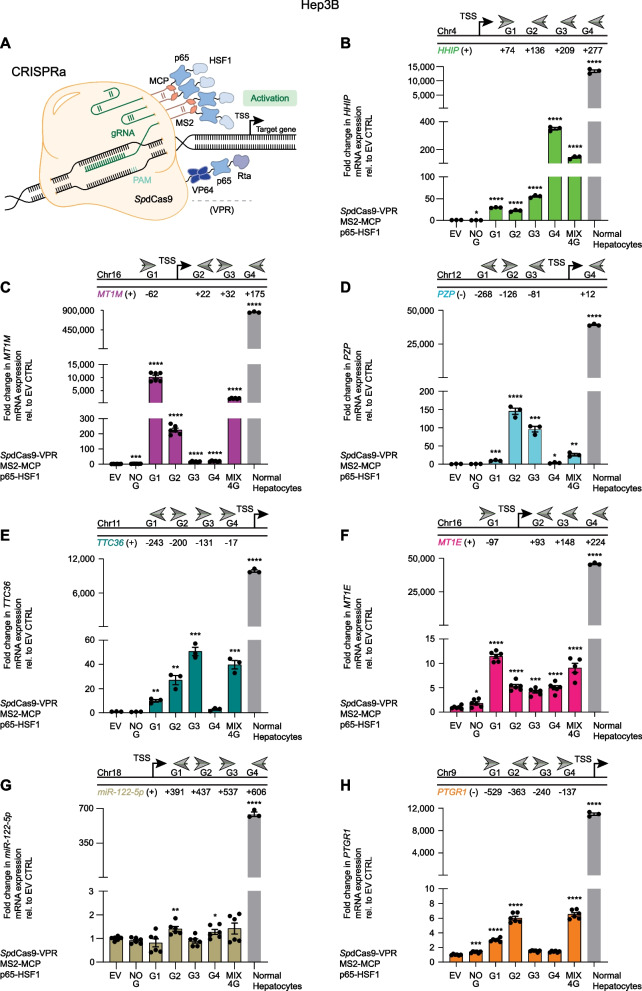
Fig. 3.
Upregulation of tumor suppressor genes by CRISPRa in Hep3B HCC cells. A Schematic representation of CRISPRa consisting of SpdCas9 C-terminally fused to the tripartite transactivator VPR (VP64, p65, and Rta) and coupled with the gRNA-MS2-MCP system that recruits the bipartite transactivator p65-HSF1 for targeted epigenetic editing. As indicated for each tumor suppressor gene (TSG), gRNAs, designated as G1, G2, G3, and G4, direct CRISPRa to the forward (right arrow) or reverse (left arrow) DNA strand within the regulatory region and proximal promoter of the TSG. gRNA numbering (±) refers to the distance in base pairs from the transcription start site (TSS) of each targeted TSG. B–H Reactivation of TSGs by CRISPRa was evaluated by qRT-PCR 48 h after transient transfection. Fold change in TSG mRNA expression from transfected cells with CRISPRa and TSG-targeting gRNAs, and normal hepatocytes was normalized to control transfections with empty vector (EV) and compared to CRISPRa with no gRNA (NO G) for statistical analysis. From left to right: B HHIP: *P = 0.0390, ****P < 0.0001; C MT1M: ***P = 0.0004, ****P < 0.0001; D PZP: ***P = 0.0006, ****P < 0.0001, ***P = 0.0002, *P = 0.0359, **P = 0.0013, ****P < 0.0001; E TTC36: **P = 0.0013, **P = 0.0022, ***P = 0.0001, ***P = 0.0004, ****P < 0.0001; F MT1E: *P = 0.0349, ****P < 0.0001, ***P = 0.0002; G miR-122-5p: **P = 0.0014, *P = 0.0119, ****P < 0.0001; H PTGR1: ***P = 0.0001, ****P < 0.0001. Data presented as means ± SEM (n = 3), and P values were determined by unpaired t-test. SpdCas9 Streptococcus pyogenes deactivated Cas9 protein adopted for epigenome engineering, MS2 RNA aptamer, MCP MS2 coat protein, HSF1 heat shock factor 1, Chr chromosome, (+) forward DNA strand, (−) reverse DNA strand, and MIX 4G combination of all four gRNAs targeting a TSG